Table 1 Estimates of nucleotide diversity in mosquitoes (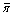 ), obtained from different source populations, numbers of loci sequenced (N loci) and sample sizes (N).
), obtained from different source populations, numbers of loci sequenced (N loci) and sample sizes (N).
Species | Source of sequenced samples | N loci | N |
| Study |
|---|---|---|---|---|---|
An. gambiae ss | mixed wild population (M) | 8 | 203 | 0.0208 | [30] |
mixed wild population (M) | 142 | 203 | 0.0043 | [30] | |
single wild population (M) | 109 | 8 | 0.0076 | [10] | |
mixed wild populations (M) | 660 | 104 | 0.0079 | Present study | |
3 lab strains (M) | 35 | 7–9 | 0.0091 | [11] | |
mixed wild populations (S) | 8 | 223 | 0.0191 | [30] | |
mixed wild populations (S) | 142 | 223 | 0.0043 | [30] | |
mixed wild population (S) | 109 | 9 | 0.0092 | [10] | |
mixed wild populations (S) | 660 | 104 | 0.0082 | Present study | |
An. arabiensis | single wild population | 22 | 23 | 0.0040 | [30] |
lab population1 | 109 | 8 | 0.0064 | [10] | |
An. funestus | single wild population | 50 | 21 | 0.0079 | [31] |
Ae. aegypti | 3 lab strains | 25 | n/a | 0.0122 | [32] |
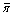 ), obtained from different source populations, numbers of loci sequenced (N loci) and sample sizes (N).
), obtained from different source populations, numbers of loci sequenced (N loci) and sample sizes (N).