Figure 5
From: ChIP-chip versus ChIP-seq: Lessons for experimental design and data analysis
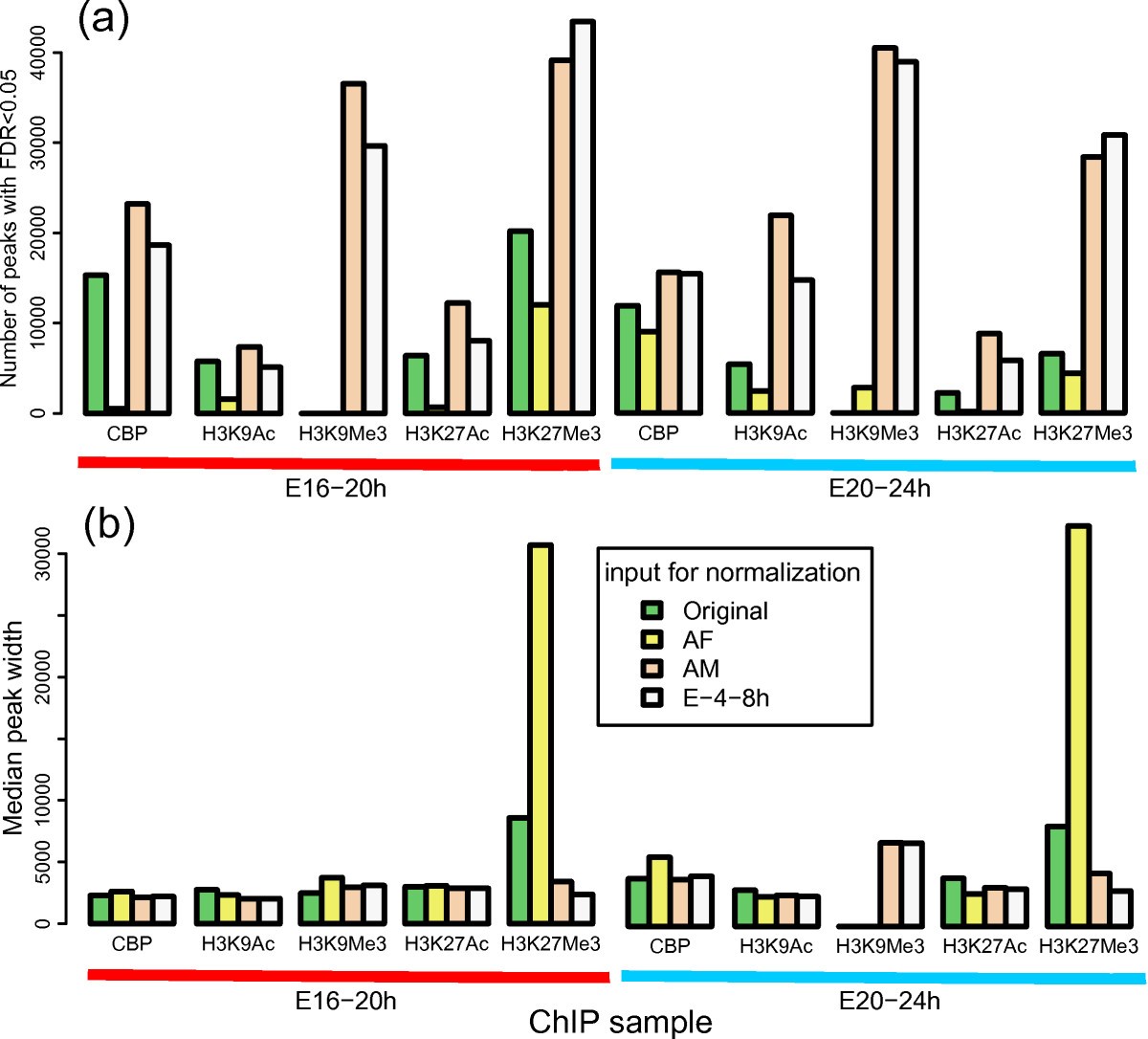
Effect of normalization with different INPUT-seq on ChIP-seq peak calling. We compared the number of peaks (a) and median peak width (b) of 10 ChIP-seq samples (CBP, H3K9Ac, H3K9Me3, H3K27Ac, H3K27Me3 at E16-20 h and E20-24 h) where each of them was normalization against four different input DNA samples (the input for from the matching time point, AdultFemale, AdultMale, and E-4-8 h). Peak calling was performed with SPP using the same parameters. Clearly peak detection is significantly affected by using different input DNA library as background control. In general, more peaks are identified as statistically significant (FDR < 0.05) when normalized with an INPUT-seq library with higher sequencing depth, although the magnitude of the differences vary across different ChIP datasets.