Figure 6
From: Comparative analysis of sequence features involved in the recognition of tandem splice sites
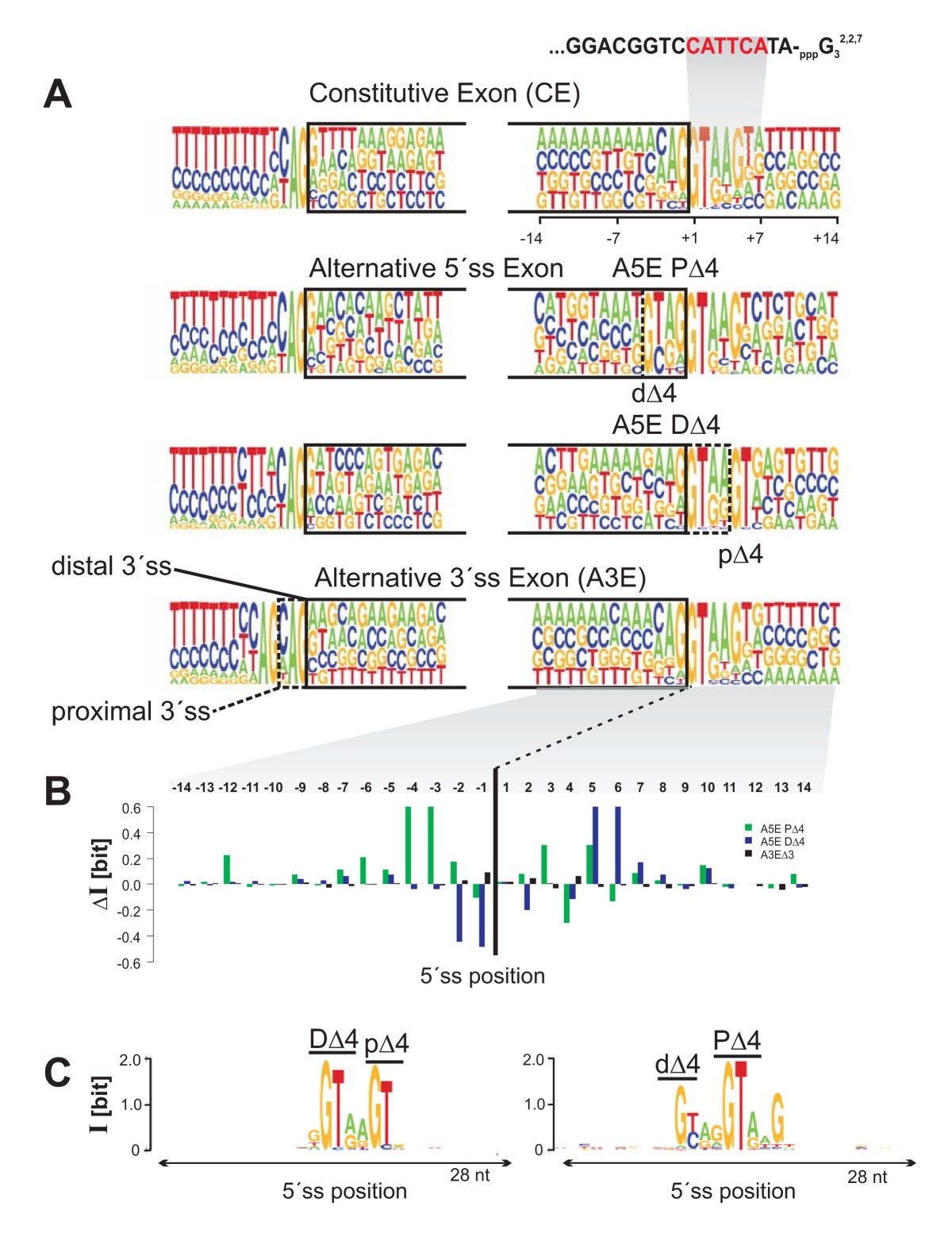
Splice site signals and sequence conservation around splice sites. A) Pictograms of 5'ss and 3'ss of constitutive, PΔ4 and DΔ4, and A3EΔ3 splicing exons. The height of a nucleotide represents the frequency of occurrence at a given position, represented in the range of 14 nucleotides around the splice junctions. Above the constitutive 5'ss, the 3'-end of the U1 snRNA is indicated. B) Information score difference (ΔI) between PΔ4 and DΔ4, respectively, and constitutive splicing exons, as well as A3EΔ3 and constitutive splicing exons. For each position, ΔI > 0 (ΔI < 0), indicates more (lack of) information of an alternative compared to a constitutive splice site. C) Sequence conservation of human PΔ4 and DΔ4 splice sites and splice sites of exons of orthologous mouse genes, 'anchored' at major splice sites and with > 80% exon sequence identity.