Fig. 1
From: 7C: Computational Chromosome Conformation Capture by Correlation of ChIP-seq at CTCF motifs
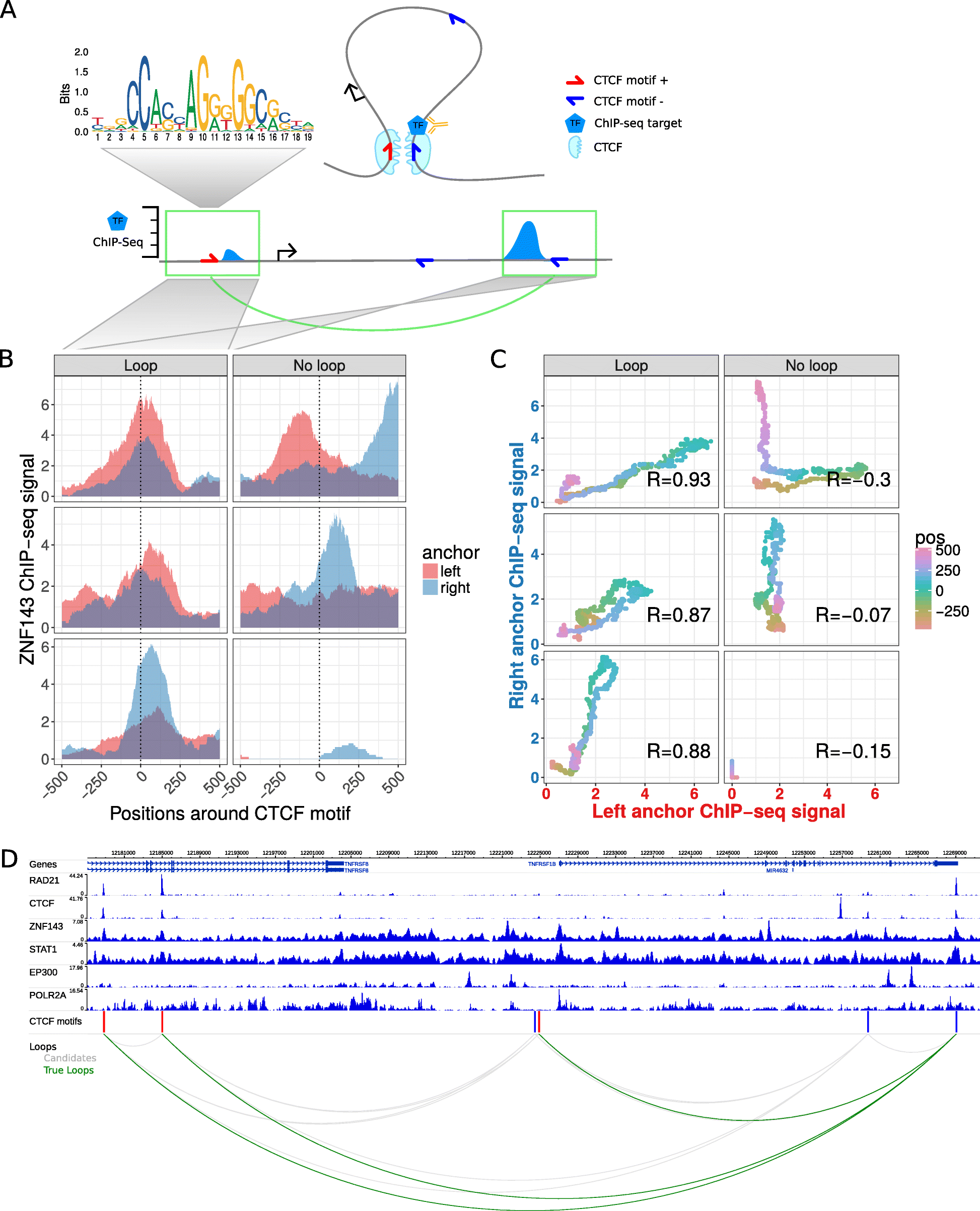
Chromatin looping interactions result in ChIP-seq coverage signals at direct and indirect bound loop anchors. a Schematic illustration of a chromatin loop with CTCF motifs at the loop anchors (top right). A transcription factor (TF) binds directly at the right loop anchor close to the CTCF motif. This results in a ChIP-seq coverage peak at the directly bound locus (bottom right) and in a minor signal at the other loop anchor (bottom left), both at the same distance to each CTCF motif. b Znf143 ChIP-seq coverage at six selected example CTCF motif pairs of which the ones in the left panel interact via loops according to Hi-C and ChIA-PET data and the ones in the right panel do not interact. The ChIP-seq coverage signal for each loci pair is shown in red for the left anchor region and in blue for the right anchor region, according to the distance to the CTCF motif (x-axis). Interacting CTCF motif pairs show more similar ChIP-seq coverage signals, which are often enriched at similar distances to the CTCF motif pairs, while the profiles of non-interacting pairs are less similar. c The similarity of ChIP-seq profiles by correlation of the ChIP-seq coverage signals of the selected motif pairs in (b). For each pair, the coverage at the right anchor is plotted versus the coverage at the left anchor at the same distance (color coded) from each CTCF motif. The Pearson correlation coefficient (R) of the dots is higher for interacting loci pairs. d Example loci on chromosome 1 shown in the genome-browser with six ChIP-seq tracks. Red and blue bars indicate CTCF recognition motifs on the forward and reverse strand, respectively. The bottom panel shows CTCF motif pairs in gray (candidates) and actually interacting pairs in green, according to ChIA-PET and Hi-C data