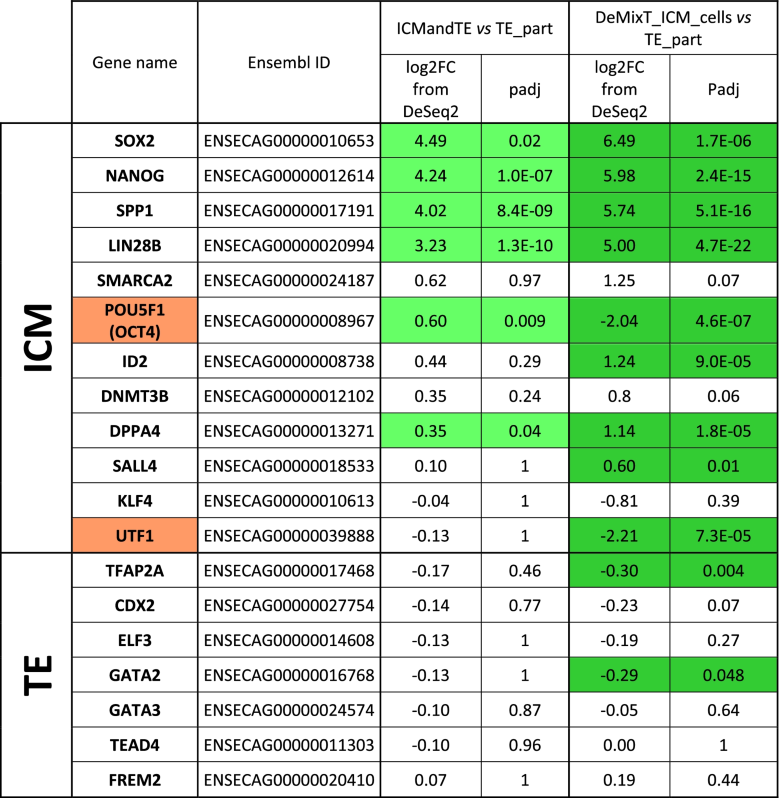
- Gene expressions were obtained from RNA of 11 equine embryos bissected in two hemi-embryos: one part is composed only of trophoblast (TE), TE_part, while the other part is composed of TE and inner cell mass (ICM), ICMandTE. As it is impossible to estimate the proportion of each cells in ICMandTE, deconvolution algorithm (package DeMixT) was used to estimate gene expression of these different kind of cells. DeMixT_ICM_cells dataset is the deconvoluted gene expression of ICM cells from ICMandTE. Log2 fold change (log2FC) and padj (adjusted p-value with Benjamini-Hochberg correction) were obtained with Deseq2 package. TE_part is the reference group in both analyses: when log2 fold changes (log2FC) > 0, gene is more expressed in ICMandTE or DeMixT_ICM_cells while when log2FC < 0, gene is more expressed in TE_part. Green is used to represent gene differentially expressed in the present study. Orange is used to represent gens that have been previously identified as predominant in the ICM [37] but which are identified here as predominant in the TE

