Fig. 5
From: New insights into the genome of Rhodococcus ruber strain Chol-4
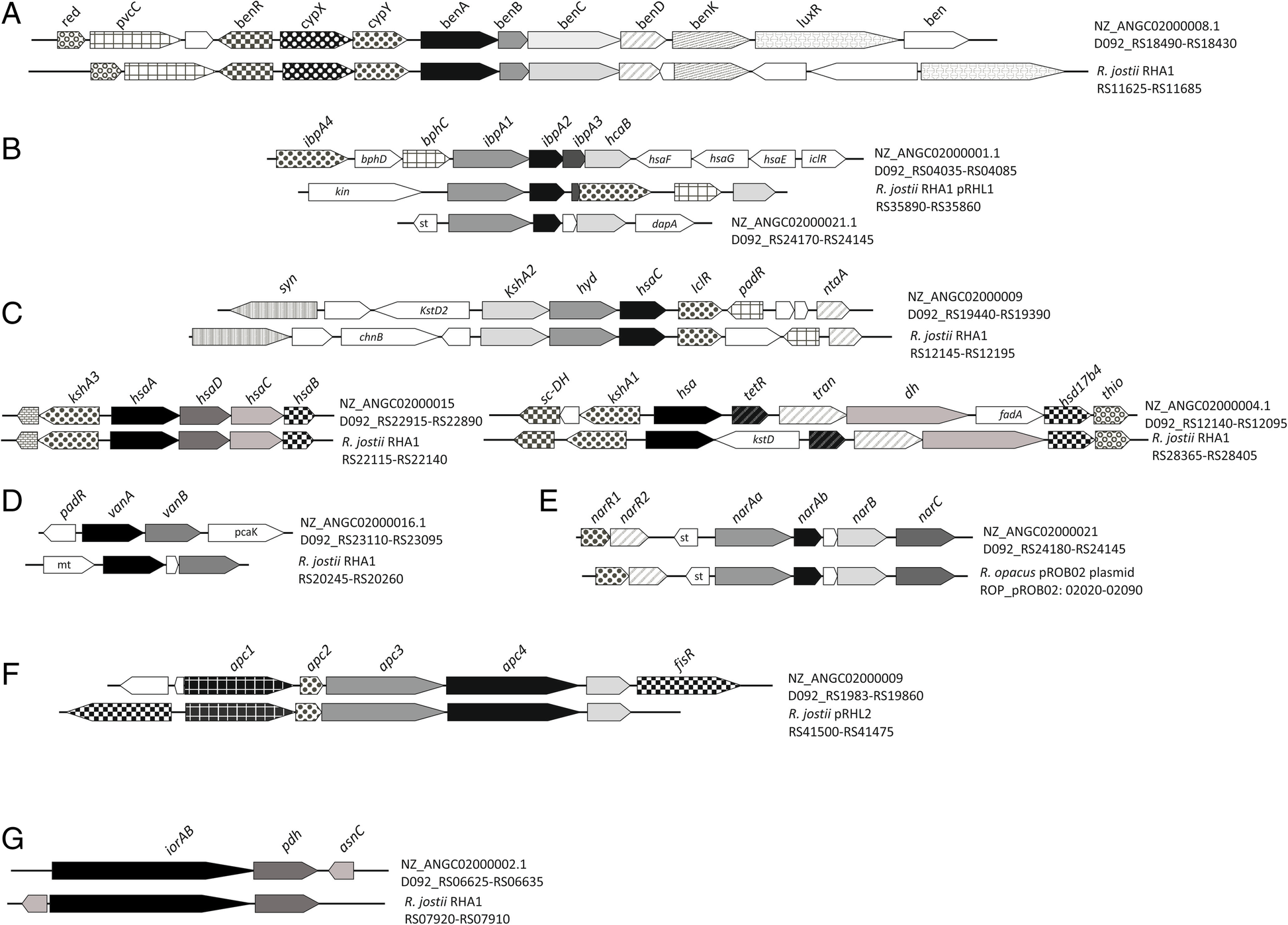
Peripherical routes in R. ruber. Abbreviations: a Benzoate degradation: red: flavin reductase; pvcC: pyoverdin chromophore biosynthetic protein; benR: transcriptional regulator (AraC family); cypX: cytochrome P450 monooxygenase; cypY: putative phenol hydroxylase; benA: benzoate 1,2-dioxygenase α subunit; benB: benzoate 1,2-dioxygenase β subunit; benC: benzoate dioxygenase, ferredoxin reductase component/1,2-dihydroxycyclohexa-3,5-diene-1-carboxylate dehydrogenase; benD: 1,2-dihydroxycyclohexa-3,5-diene-1-carboxylate dehydrogenase; benK: benzoate MFS transporter; luxR: transcriptional regulator (luxR family) putative.; ben: benzoate transport protein. b Isopropylbenzene pathway: ipbA4: ferredoxin reductase; bphD: 2-hydroxy-6-oxo-2,4-heptadienoate hydrolase; bphC: 2,3-dihydroxybiphenyl 1,2-dioxygenase; ipbA1: isopropylbenzene 2,3-dioxygenase or IPB-dioxygenase, ISP large subunit; ipbA2: IPB-dioxygenase (ISP small subunit); ipbA3: IPB-dioxygenase ferredoxin; hcaB: hydroxybenzaldehyde dehydrogenase; hsaF, hsaG; hsaE: previously described (Fig. 4); iclR: transcriptional regulator (IclR family); kin: sensor kinase; st: sterol-binding domain protein; dapA: 4-hydroxy-tetrahydrodipicolinate synthase. c Steroids pathway: syn: non-ribosomal peptide synthetase; kstD: 3-oxosteroid 1-dehydrogenase; kshA: ketosteroid-9-α-hydroxylase, oxygenase; hyd: hydroxylase; hsaC: 2,3-dihydroxybiphenyl 1,2-dioxygenase; iclR: transcriptional regulator (IclR family); padR: transcriptional regulator (PadR family); ntaA: nitrilotriacetate monooxygenase component B; chnB: cyclohexanone monooxygenase; hsaA: flavin-dependent monooxygenase; hsaD: 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase; hsaC: iron-dependent extradiol dioxygenase; hsaB: flavin-dependent monooxygenase reductase subunit; sc-DH: short-chain dehydrogenase; hsa: monooxygenase; tetR: probable transcriptional regulator (TetR family); tran: acetyl-CoA acetyltransferase; dh: acyl-CoA dehydrogenase; fadA: 3-ketoacyl-CoA thiolase; hsd17b4: 3-α,7-α,12-α-trihydroxy-5-β-cholest-24-enoyl-CoA hydratase; thio: thioesterase. d Vanillate: padR: transcriptional regulator (PadR family); vanA: vanillate o-demethylase oxygenase subunit, flavodoxin reductases (ferredoxin-NADPH reductases) family 1; vanB: vanillate o-demethylase oxidoreductase; pcaK: 4-hydroxybenzoate transporter; mt:methyltransferase. e Naphtalen. (nar genes: R. opacus plasmid pROB02:AP011117): narR1 and narR2: putative naphthalene degradation regulatory protein; narAa: nidA, naphthalene dioxygenase large subunit; narAb: (nidB) naphthalene dioxygenase small subunit; narB: (nidC) 1,2-dihydro-1,2-dihydroxynaphthalene dehydrogenase; narC: (nidD) putative aldolase NarC. f Acetophenone carboxylase (anaerobic): apc1–4: acetophenone carboxylase subunits; fisR: transcriptional regulato (Fis family). g Aminoacid: iorAB: indolepyruvate ferredoxin oxidoreductase (α and β subunits); pdh: glutamate / leucine / phenylalanine / valine dehydrogenase; asnC: transcriptional regulator (AsnC family). All R. jostii RHA1 genes have the prefix “RHA1_” not included in the figure