Fig. 1
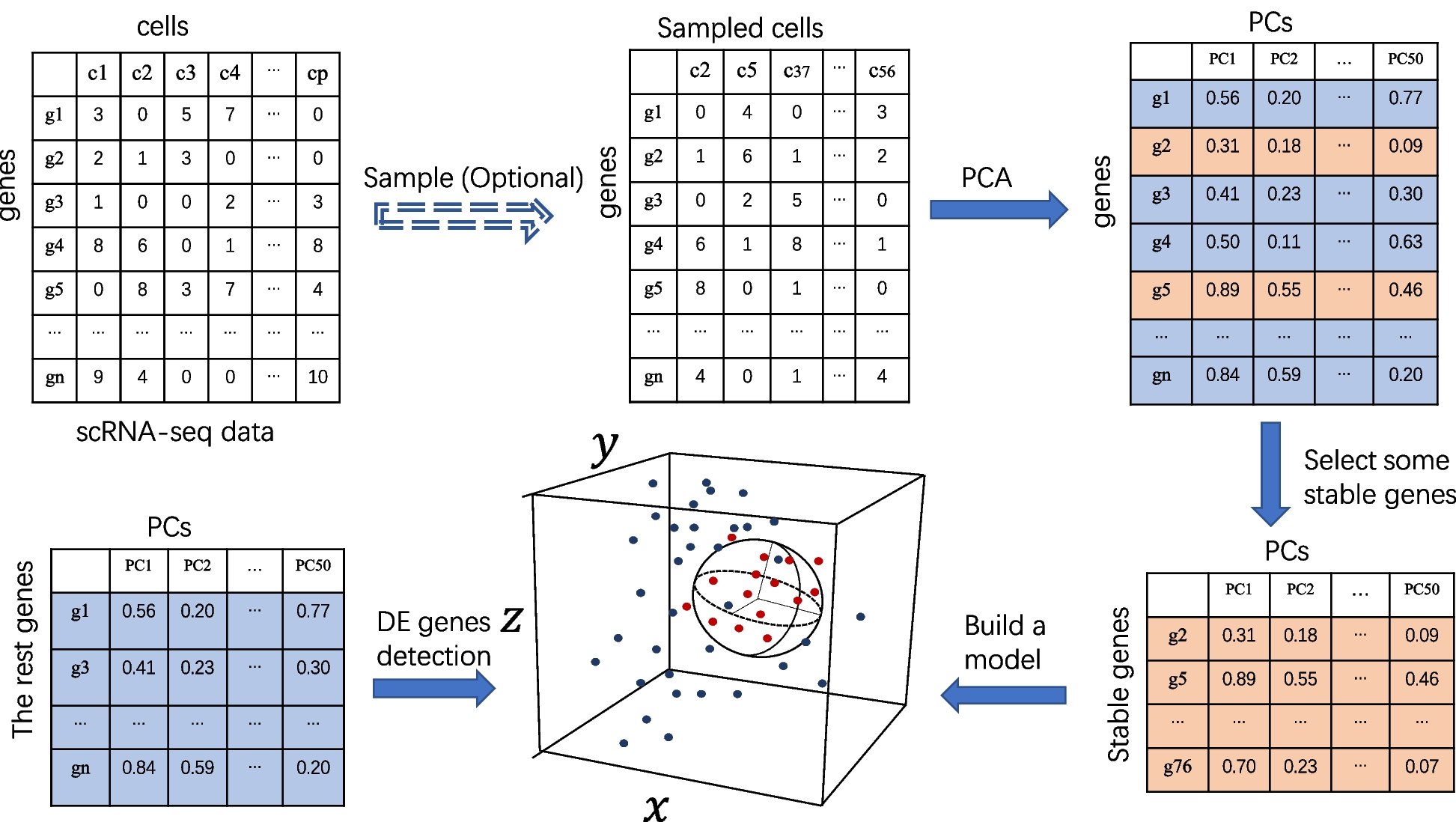
Overview of the scMEB workflow. Randomly sampling some important cells before PCA is optional, and the purpose is to speed up the calculation. The first 50 PCs could be obtained from the sampled/unsampled cells. A small part of the stable genes was used to build the model. A sphere that encloses the training data in the feature space was found. The remaining genes were divided into DEGs or non-DEGs according to whether they were outside or inside the enclosing ball, respectively