- Research article
- Open access
- Published:
Comparative genome-wide characterization leading to simple sequence repeat marker development for Nicotiana
BMC Genomics volume 19, Article number: 500 (2018)
Abstract
Background
Simple sequence repeats (SSRs) are tandem repeats of DNA that have been used to develop robust genetic markers. These molecular markers are powerful tools for basic and applied studies such as molecular breeding. In the model plants in Nicotiana genus e.g. N. benthamiana, a comprehensive assessment of SSR content has become possible now because several Nicotiana genomes have been sequenced. We conducted a genome-wide SSR characterization and marker development across seven Nicotiana genomes.
Results
Here, we initially characterized 2,483,032 SSRs (repeat units of 1–10 bp) from seven genomic sequences of Nicotiana and developed SSR markers using the GMATA® software package. Of investigated repeat units, mono-, di- and tri-nucleotide SSRs account for 98% of all SSRs in Nicotiana. More complex SSR motifs, although rare, are highly variable between Nicotiana genomes. A total of 1,224,048 non-redundant Nicotiana (NIX) markers were developed, of which 99.98% are novel. An efficient and uniform genotyping protocol for NIX markers was developed and validated. We created a web-based database of NIX marker information including amplicon sizes of alleles in each genome for downloading and online analysis.
Conclusions
The present work constitutes the first deep characterization of SSRs in seven genomes of Nicotiana, and the development of NIX markers for these SSRs. Our online marker database and an efficient genotyping protocol facilitate the application of these markers. The NIX markers greatly expand Nicotiana marker resources, thus providing a useful tool for future research and breeding. We demonstrate a novel protocol for SSR marker development and utilization at the whole genome scale that can be applied to any lineage of organisms.
The Tobacco Markers & Primers Database (TMPD) is available at http://biodb.sdau.edu.cn/tmpd/index.html
Background
Simple sequence repeats (SSRs), otherwise known as short tandem repeats (STRs) or microsatellites, are abundant and broadly distributed in eukaryotic and prokaryotic genomes. The length of an SSR shows extensive intra- and interspecific variation, primarily because of high rates of DNA replication error within SSRs [1,2,3]. Hence, SSRs are widely used for designing PCR-based markers that can be very useful for population genetic characterizations, genome mapping, and such applications as tagging trait-associated genes during marker-assisted selection [4]. The genomic quantity and distribution of SSRs differs between dicot and monocot plants [5]. Analyses of SSR distribution revealed that di-nucleotide repeats are more common than tri-nucleotide repeats in dicots, and the most frequent motifs are AT and ATT/AAT [6]. In monocot grass genomes, the most abundant motif is GA/TC, the A/T monomer and then the GCG/CGC trimer [7]. Recent analyses found that the SSR abundance increases linearly with genome size in fully assembled grass genomes [7]. Most SSRs are in intergenic regions and in 5’-UTRs [5, 6]. Characterization of the SSRs across an entire genome provides the foundation for comprehensive SSR marker development. In fact, genome-wide SSR markers have been designed for several plant species after their genome sequences were released, including in rice [8], soybean [9], Brachypodium [10], maize [11], foxtail millet [12], Brassica [13] and cotton [14].
Nicotiana, a member of the Solanaceae family, is one of the most important research model plants and is of high agricultural value worldwide [15, 16]. Species in the genus Nicotiana have large genome sizes, averaging ~ 2.5 Gb for diploids and ~ 4.5 Gb for tetraploids, which made it challenging to sequence whole genomes [17]. With recent advances in next generation sequencing (NGS) technologies, extensive genome sequence assemblies were generated for seven Nicotiana species or varieties and made available to the public [17]. These sequences are derived from the diploids N. otophora (N. oto), N. sylvestris (N. syl) and N. tomentosiformis (N. tom) and from four allotetraploids, namely N. benthamiana (N. ben) and three major commercial varieties of N. tabacum (N. tab): TN90, K326, and BX [18,19,20,21]. The N. syl and N. tom lineages diverged about 15 million years ago [22]. As confirmed by genomic sequences [20], N. syl and N. tom are the ancestors of N. tab. An interspecific hybridization between N. syl and N. tom formed N. Tab. 0.2 million ago [23]. The three N. tab varieties are nearly identical [20]. N. ben is a tetraploid (38 chromosomes) that formed ~ 10 million years ago so it is far distinct from tetraploid N. Tab (48 chromosomes) [17,18,19,20,21]. Although these genomic sequences are only assembled into contigs and scaffolds, they are sufficiently long in genic regions to allow characterization of SSRs at a whole-genome scale.
Genetic markers have been used in many types of biological research, including the construction of genetic maps, population genotyping, phylogenetics, genome comparisons, gene mapping, quantitative trait loci analysis and marker-assisted breeding [4, 20, 24, 25]. Single nucleotide polymorphisms (SNPs) and SSR markers are the two types of genetic markers most commonly employed nowadays [24]. The most reported markers in Nicotiana currently are SSR markers. Two sets of SSR markers, 5119 named PT markers and 4886 named TM or TME markers, were designed from N. tab partial genomic DNA or EST sequences by Bindler and colleagues, and by Tong and coworkers, respectively [26, 27]. Of those SSR markers, 2318 PT SSR markers were anchored in 24 linkage groups of a genetic map of tetraploid N. tab [27], while 590 SSR markers were placed in another N. tab genetic map [26]. In addition, markers were developed and used for the diploid N. tom map, involving 489 SSRs and conserved ortholog set II (COS II) markers. The COS II markers are gene sequences of a small number of orthologous loci across the investigated species [28]. Similarly the genetic map of the diploid N. acuminata contains a mixture of 308 SSRs and COS II markers [28]. Some of the SSR markers used in the diploid genetic maps are redundant with some of the PT markers.
For the species of Nicotiana, all with large genomes, the currently available markers represent only a small part of all possible SSR loci, and these may not be sufficient for some marker-based applications. In addition, most reported SSR markers were developed for one or two particular species or varieties only. Hence, the development of a whole-genome set of SSR markers across multiple species of Nicotiana genus is warranted. One complicating issue is that many Nicotiana species are polyploid, which can make it difficult to associate any given set of allelic polymorphisms with a particular homoeologous genome. Therefore, the study of markers in multiple genomes of Nicotiana will be most helpful if it provides exact allele information. Cross-species marker transferability has also proven to be an important issue during marker development in multiple plant lineages [29]. With the availability of multiple genome sequences of Nicotiana species, analysis of both the homoeology and transferability of SSRs markers among species is more feasible, as previously shown [28].
So far, genome-wide SSR distribution has not been well investigated across Nicotiana species. Our study discovered and characterized a comprehensive set of SSRs in silico in seven sequenced Nicotiana genomes mentioned above. We used our novel software package called GMATA® [7] to facilitate comparative SSR analyses of these large Nicotiana genomes, and also developed an efficient protocol for SSR marker development that can be employed across any genome. The discovered Nicotiana SSRs and developed SSR markers were used to investigate marker polymorphism in these seven species. We also developed an online database called the Tobacco Markers & Primers Database (TMPD) to facilitate the use of the SSR information and markers.
Results
Distributions of SSR types, lengths and locations in Nicotiana genomes
We analyzed perfect repeated SSRs with repeat unit length between 1 and 10 bp across seven publicly available Nicotiana genome sequences, including three diploids and four tetraploids species. The sequences used in this study total ~ 20 Gb, and are from N. ben, N. syl, N. tom, and N. oto, and from N. tab cultivars TN90, K326, and BX. These sequences represent a respective ~ 74%, ~ 80%, ~ 78%, ~ 81%, ~ 84%, and ~ 73% of each of the genomes, and are primarily deficient in the highly repetitive regions such as nested transposon blocks that tend to be poor (< 1%) in SSRs [14, 30, 31]. We identified 3010,76 such SSR loci in these seven Nicotiana genomes using the software GMATA [7]. The average density of SSRs in the DNA sequence is ~ 155 SSRs/Mb, or one SSR for every ~ 6 kb (Table 1). The properties of SSR occurrence, relative frequency and distribution of each type in these Nicotiana genomes were compared in Additional file 1: Table S1.
The types of repeated units in all identified SSRs were characterized. Mono-, di- and tri-nucleotide repeats are the major types of SSRs, accounting for > 98% (Fig. 1). Di-nucleotide repeats are the most frequent SSR type in Nicotiana, with a frequency of 61–65% (Fig. 1). Both mono- and tri-nucleotide repeats account for 10–20%, and tetra-nucleotide SSRs comprise about 0.9–1.7% (Fig. 1). The remaining SSR types, from penta-nucleotide to deca-nucleotide, together provide less than 1% of the total SSRs in all investigated Nicotiana genomes. However, given these large data sets, this < 1% provides 11,009 long-unit SSR loci that we have discovered in Nicotiana. Interestingly, deca-nucleotide SSRs were detected in all investigated Nicotiana except N. tom. Comparison across seven genomes revealed that six of seven Nicotiana genomes have the same abundance rank of di-, mono-, tri- and tetra-nucleotide SSRs while N. ben has the order of di-, tri-, mono-, and tetra-nucleotide. The remaining SSR types exhibit different ranks in each species (Additional file 1: Table S1).
SSR polymorphism was then determined by in silico characterization of SSR length variation across the seven Nicotiana genomes (Additional file 1: Table S1). SSR length was classified into two categories in accordance with previous designations [32]. The class II type (length < 20 bp) is more abundant than the class I type (length > = 20 bp) in Nicotiana. Class II type SSRs account for approximately 70, 68 and 72% of all SSRs in N. ben, N. tom, N. oto, respectively, while there is a higher but very similar percent of ~ 75% in N. syl and the three N. tab varieties TN90, K326, BX. The similarity of SSR length in the three N. tab varieties is consistent with the overall similarity of these genomes [20]. The longer class I SSRs have a greater chance of hyper-polymorphism [32]; therefore, length is an important factor for marker development. The frequency of class I SSRs in Nicotiana genomes ranges from 23 to 32%. N. tom has a higher percentage (~ 32%) of class I SSRs than does N. syl and N. tab. There is a similar frequency (~ 25%) of class I SSRs in N. syl and N. tab. The overall trend of SSR length distribution is that the frequency of occurrence decreases as the SSR length increases (Additional file 2: Figure S1). The rank of SSR abundance in both classes is different between investigated Nicotiana varieties and species. N. ben and N. tom share a very similar abundance order of SSR length when SSR length is < 40 bp, and have species-specific order for SSRs > 40 bp.
To investigate the SSR locations in genomic sequences, we extracted the top five longest assemblies (~ 800 kb) from seven genomic assemblies and compared the location of SSRs in these scaffolds. The results showed that the location of SSRs did not follow any obvious pattern in linear genomic sequences (Fig. 2). We further compared the SSR distribution between gene coding sequences (CDS) and all genomic sequences. The results showed that most (99.1%) of the SSR are in non-genic regions, and the longer repeated units of 7–10 bp are represented only in non-genic sequences. The most abundant motif is AT/AT and TA/TA in non-genic sequence while it is TC/GA in CDS (Table 2).
The locations of SSR in Nicotiana genome sequences. Plots show SSR locations in five large scaffolds of Nicotiana genome sequence. The top five largest sequences were extracted from seven Nicotiana genome assemblies, and SSR positions were mined using software GMATA®. The gi number shows the genome sequence ID at GenBank
SSR motifs in Nicotiana
A detailed characterization of motifs in Nicotiana SSRs was carried out using the GMATA software [7]. We grouped the unit motifs into pairs if sequences of two motifs were found to be complementary, because of the unknown orientation of DNA strands, which may lead to different encoded frames, in the current draft genome assemblies. There are 817, 631, 639, 768, 879, 844, and 847 types of grouped motif pairs identified in N. ben, N. syl, N. tom, N. oto, and N. tab TN90, K326, BX, respectively. Generally, the abundance of grouped motif pairs in Nicotiana increased with the size of the genome sequence. N. tab has the highest number of grouped motif pairs, but this number differs somewhat between N. tab varieties. The motif pairs ranking at the 20 highest frequencies in each of the seven Nicotiana genomes are shown in Additional file 1: Table S1. Motif pairs AT/AT are predominant, accounting for 44–50% of total SSRs in the seven examined Nicotiana genomes. Grouped motif pairs GC/GC and CG/CG are the rarest di-nucleotide motifs, at frequencies of ~ 0.1–0.17%, ranking after twenty tri-nucleotide motif pairs. The distribution of tri-nucleotide motif pairs displayed major genetic variation among these Nicotiana genomes. Motif pairs CAA/TTG and GTT/AAC are the most abundant tri-nucleotides, with a total frequency of ~ 5% in six of the genomes, but N. tom has more motif pair ATT/AAT than motif pair CAA/TTG and GTT/AAC. AAAT/ATTT is the most abundant tetra-nucleotide motif pair in six of the genomes, but is the second most abundant in N. ben, after the ATAC/GTAT motif pair. The remaining motif pairs with repeat lengths > 4 bp are the rarest motifs (Additional file 1: Table S1).
Genome-wide SSR marker development in multiple Nicotiana genomes
To develop a comprehensive set of SSR markers at a genomic sequence scale in multiple Nicotiana genomes, the sequences flanking SSR loci in the seven Nicotiana genomic sequences were used to design PCR primers with the GMATA software [7]. In total, 1,943,197 loci (78.3% of the total analyzed) yielded high quality primer pairs. Of the seven Nicotiana genomes, most genomic sequences are highly similar [18, 20]; therefore, it is likely that designed primers for one genome will commonly work in another. In our approach, the same primer pairs designed from more than one genome dataset were condensed to a single unique primer pair. Further clustering was conducted to keep unique primer pairs based on identity of the primer’s sequence. Each unique SSR primer pair was then considered as a potential marker and assigned a unique marker ID. In total, 1,224,048 unique SSR markers were designed. We call these NIX (Nicotiana multiple (X) genome) markers. The information on all designed NIX markers is provided in “sts” format, according to the NCBI’s rule for marker information, and is freely available to the public in our online database TMPD as described later.
To investigate the novelty of the NIX markers, a comparison was conducted between NIX markers and previously published markers in Nicotiana. After removing redundancy of published markers based on primer sequence similarity, 5397 unique SSR markers from Bindler et al. [27, 33] and 387 unique markers from Wu et al. [28, 34] were classified as reported markers. Of these, 2143 (37.0%) of the reported markers exhibited only one primer that overlapped with those in NIX markers. A total of 264 of the NIX markers yielded overlapping primers for both members of a pair, and are thus identified as duplications of published marker pairs, indicating that 99.98% of NIX makers are novel (Additional file 2: Figure S1). However, a few novel NIX markers do amplify the same SSR loci published in earlier studies [27, 28, 33, 34].
In silico analysis of NIX marker polymorphism across genomes
For marker utilization, it is vital to know the amplification efficiency, allele size, and level of polymorphism (i.e., number of alleles). Thus, an in silico simulated PCR was carried out to investigate polymorphism across the seven Nicotiana genomic sequences. Two copies of genes in tetraploid Nicotiana will produce two alleles of PCR product if repeat numbers are different. We found that all the designed NIX markers would produce at least one amplicon in at least one investigated genome. In the diploids, ~ 30% of designed NIX markers would amplify DNA in N. syl while a lower percentage (~ 25%) would provide an amplification product in both N. tom and N. oto. For tetraploids, ~ 53% of NIX markers are amplifiable in each of the three N. tab varieties, while only ~ 23% would amplify an SSR product in N. ben. Because the information regarding common and species-specific NIX markers will be extremely useful for marker selection during application, we checked this for all NIX markers. A total of 9948 NIX markers (~ 0.8%) will be amplifiable in all seven investigated Nicotiana species or varieties, so we infer these markers are very likely to be amplifiable in any other Nicotiana species. There are also many species-specific NIX markers (e.g., 128,639 such markers in N. tab). N. ben has more species-specific NIX markers than any other, which may be explained by the fact that N. ben is the most distantly related of the Nicotiana species analyzed [19]. The fewest NIX markers were found in N. syl (Fig. 3a). Of the NIX markers in the three N. tab varieties, 566,858 (78.6%) are in common, while only a few markers (2.3–3.5%) are variety-specific (Fig. 3b). Despite the low divergence between the N. tab varieties, only ~ 3.6% (28,416) of the NIX markers were shared with both progenitors N. syl and N. tom. Besides the common NIX markers, N. tab appears to have inherited a different number of NIX markers from its two putative progenitors, 34.8% from N. syl and 25.6% from N. tom (Fig. 3c).
Venn diagram showing the number of common and specific NIX markers in Nicotiana species and varieties (a), varieties of N. tab (b), N. tab K326 and putative progenitors (c). N. ben, N. syl, N. tom, N. oto, N. tab TN90, K326 and BX represent N. benthamiana, N. sylvestris, N. tomentosiformis, N. otophora, N. tabacum TN90, N. tabacum K326, and N. tabacum BX, respectively
A total of 744,993 (61%), 211,845 (17%) and 106,472 (8.7%) of the designed NIX markers are predicted to produce a respective one, two or three PCR fragments from the combined sequences of these seven genomes. Some NIX markers will amplify only from one or a few Nicotiana genomes. Of the amplifiable NIX markers in a given genome, on average, ~ 81% of the NIX markers are predicted to produce one fragment while ~ 8% will produce two fragments. Species N. ben has the lowest percent (~ 6%) of NIX markers producing two fragments while N. oto has the highest percent (~ 11%). In the seven Nicotiana genomes combined, ~ 3.8% of NIX markers yield many predicted fragments (> = 11), suggesting that they may be amplified from repeated DNA sequences.
For all 1,224,048 NIX markers, the predicted amplicons were scored for predicted length polymorphisms across the seven genomic sequences (Table 3). A total of 430,848 markers (35.2%) showed polymorphism between at least two of the investigated Nicotiana genomes. The polymorphism information is available in the TMPD database.
Experimental validation of SSR markers
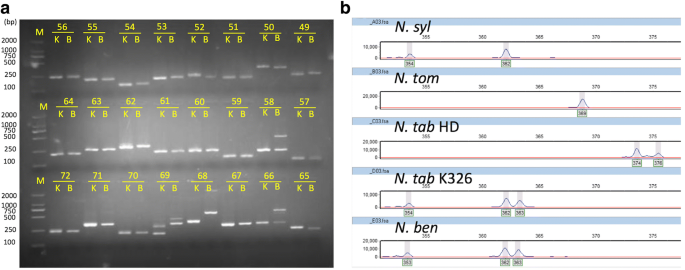
In total, 120 NIX markers were randomly selected from the predicted commonly amplifiable markers for experimental validation by PCR amplification on a testing panel of five Nicotiana accessions, including four Nicotiana species of N. syl, N. tom. N. Tab K326 and N. ben with known genomic sequence and additional species N. glutinosa or accession N. Tab HD with unknown genomic sequence (Additional file 3: Table S2). Of the tested NIX markers, 100% produced amplicons from at least four of the entire panel (Fig. 4a, Additional file 3: Table S2). For each of the Nicotiana accession, more than 99% of the markers produced amplicons in the expected size range. High amplification rates of these markers in the species N. glutinosa or accession N. Tab HD may indicate a high level of usefulness in other Nicotiana species or accessions. For most of the tested NIX markers, it is difficult to score the polymorphism among accessions in an agarose gel; however, most of the predicted polymorphisms were easily identified by fluorescent genotyping using the ABI3730X DNA fragment analyzer (Fig. 4b). We further tested the polymorphism of 24 markers with an ABI3730X, and found a high resolution of polymorphic bands (Fig. 4b). The validation results and gel images are available in our online database, TMPD, described later. The validation results demonstrated that Nicotiana NIX markers will be valuable tools that are applicable for multiple Nicotiana species or varieties.
Experimental validations of NIX markers by amplification and allele scoring. Image shows the PCR fragments resolved by agarose gel (a) or DNA fragment analyzer ABI3730X (b). N. syl, N. tom, N. Tab HD, N. Tab K326, and N. ben represent N. sylvestris, N. tomentosiformis, N. tabacum HD, N. tabacum K326 (K), and N. benthamiana (B), respectively. The numbers in the image represent validated IDs for markers
Online database of tobacco SSR markers
To facilitate the application of NIX markers, we constructed a freely available online database called the Tobacco Markers & Primers Database (TMPD) (http://biodb.sdau.edu.cn/tmpd/index.html, Fig. 5a). All NIX marker information, including primer sequences, fragment sizes, polymorphism levels and genomic location, can be downloaded. Searches can be done by NIX ID number, by primer sequence or by DNA sequence using our online Blast function (Fig. 5b and c). If the user has a DNA sequence, the Blast function in TMPD can find the nearest NIX markers to the sequence, and a further click will show the NIX marker information. A function for archiving and searching validated results was built into TMPD. TMPD hosts our existing marker validation results including gel images, and fragment information (Fig. 5d). In addition, TMPD can accept new validation results from any users.
The online tobacco markers and primers database TMPD. The TMPD database is freely available online at http://biodb.sdau.edu.cn/tmpd/index.html. Images a, b, c, and d show the TMPD interface, search function, Blast function and validation search function
Discussion
SSRs have been comprehensively characterized for some plant species after their genome sequences were published, including in Brachypodium [10], foxtail millet [12, 35, 36], Brassica [13], and cassava [37]. Currently, SSR DNA entries from 110 plant species with sequenced genomes are available in a plant microsatellite DNAs database (PMDBase) [38]. However, the genome-scale distribution of SSRs in the Nicotiana genus has been lacking due to the absence of a whole genome sequence. Recently, the availability of seven extensive genome sequence analyses from the Nicotiana genus have made deep investigation of SSRs possible [17]. The genomes, providing ~ 20 Gb of sequence data, include information from diploids and tetraploids that are closely related to commercial tetraploid tobacco, N. tab. This is the first report of the comprehensive identification, distribution analysis and comparison of SSRs in seven deeply sequenced genomes from the genus Nicotiana. To efficiently discover SSRs in large scale genome sequence data sets, the recently published software package GMATA® [7] was used. We discovered that mono-, di- and tri-nucleotide types of all the repeat units with lengths ranging from 1 bp to 10 bp are the most abundant. This observation in Nicotiana is consistent with the typical features of SSR distribution in dicots revealed by a comparison of 29 plant species [6] and in Brassica [5]. Grouped motifs TA/TA and AT/AT are predominant (44–50%), while GC/GC and CG/CG are the rarest di-nucleotides in all investigated Nicotiana genomes. In Nicotiana, most (> 68%) of the SSRs are short (< 20 bp), which is routinely correlated with a relatively low level of polymorphism for these markers. Our comparison of distribution revealed that SSRs in Nicotiana are mainly located in the regions between genes. This observation may partly explain the low rate (~ 4%) of polymorphism identified during Nicotiana SSR marker screening in an earlier study [39] that used genic sequences as the starting point for SSR discovery.
Previously, studies on Nicotiana SSRs were focused on a specific genome or variety. In this study, we compared SSRs between tetraploid N. tab and its ancestral diploids, between varieties of tetraploid N. tab, and between different tetraploid species. Comprehensive comparison revealed genome-specific SSRs in each of the seven Nicotiana genomes. Some of the absences of SSRs in certain accessions may result from incompleteness in the genome sequences. However, most genomic sequences that are missing in these seven assemblies are repetitive sequences that are generally deficient in SSRs [30]. Therefore, the SSRs represented here should reflect the general distribution of SSRs. Species N. Tab BX and N. ben, for instance, have incomplete genome sequences, believed to represent 74 and 73% of full genome sequence, respectively [19, 20]. However, N. Tab BX did not show a biased distribution of amplicons by in silico analysis compared with the other genomes. N. ben has the fewest NIX markers that are predicted in silico to produce two fragments, which may be caused by both the less complete sequences and perhaps by the fewer chromosomes (38) in this species, compared to the 48 chromosomes in other tetraploids. Perhaps the chromosome number reduction was associated with some DNA loss during tetraploid evolution.
Diploid N. oto has more grouped motifs than diploid N. syl, which indicates that the two genomes became very different after their lineages diverged from the same ancestor. This conclusion is also supported by genome sequence comparison of these two species [20]. In tetraploids, the three varieties of N. tab have very similar numbers of grouped motifs, indicating their limited degree of divergence. This high homology, also seen in earlier studies [20], is likely to reflect both recent common ancestry and human selection for desirable traits linked to some of these NIX markers. Regarding the types of SSRs, all investigated Nicotiana genomes have similar di-nucleotide type abundances. However, mono-nucleotide, tri-nucleotide and other higher-order repeats were found to vary a great deal among Nicotiana genomes. These long SSRs are the major differences in SSRs between these Nicotiana genomes. In bacteria, previous studies have shown that many SSRs in intergenic or upstream regions can have regulating functions [6], and this function may sometimes be true in large-genome eukaryotes. It will be interesting to investigate whether any of these differences will lead to phenotypic variation, particularly for SSRs within transcribed regions of genes or in regulatory domains of the adjacent chromatin. Regarding SSR length, N. ben and N. tom shared a very similar abundance order. The similar distribution of short SSR repeats in N. syl and N. tab may be partially explained by the fact that N. tab has retained more genomic sequence from N. syl than from N. tom [20, 22, 23]. We found that longer SSRs (> 40 bp) are usually genome-specific in Nicotiana. In conclusion, all genomes shared most of their SSR motifs while each genome has its own specific motifs, especially for larger repeat lengths and distribution frequencies.
SSR evolution can be revealed by comparison of the SSR and its flanking sequence. The NIX markers were designed from the sequences flanking SSRs, so the amplicons’ lengths in different Nicotiana genomes will provide insights into SSR evolution. SSRs in allopolyploids of N. tab showed considerable departure from the predicted additivity of their ancestral progenitors, N. syl and N. tom, which was demonstrated by the distribution of shared and specific NIX markers (Fig. 3c). There is an increased number of amplifiable NIX markers (142,612) in N. tab compared to the sum of N. syl-specific NIX marker (60,412) and N. tom-specific NIX markers (79,771). This difference may partially be caused by both the incomplete genome sequences and real biological differences. An increase in amplifiable NIX markers (net increase of 2429 or 1.7%) means increased SSRs in polyploids relative to their diploid progenitors. In contrast, SSR frequencies in Brassica species were observed to decrease after polyploidization [5]. The genome size in N. tab is less than the sum of their ancestral progenitors, N. syl and N. tom (Leitch et al. 2008). It has been previously noted that the genome reduction from the N. tom ancestor was greater than that from the maternal N. syl genome in natural N. tab [20, 22, 23]. This suggests that the extra SSRs in N. tab may be from the amplification of DNA from the N. syl-like ancestor and loss of DNA from N. tom ancestor, which is also supported by the genomic sequence comparison between species [20]. In addition, the variety-specific and species-specific NIX markers in the polyploids indicate either SSR formation or loss after the polyploidization or speciation events. Alternatively, this may reflect differential transmission of SSR polymorphisms from the ancestors.
SSR markers have been used in several genetic maps of Nicotiana species. For tetraploid N. tab, the genetic map contains around 2000 SSR markers [27, 33]. In the diploid N. acuminata and N. tom genetic maps, a mixture of 489 and 308 SSR plus COSII markers were also reported, respectively [28]. The total number of NIX markers designed here were more than one million, 99.98% of which do not overlap with previous markers in Nicotiana. Thus, the NIX markers reported here have increased, about 240-fold, the SSR marker resource for the Nicotiana community. We identified 9948 SSR markers that were informative on all seven Nicotiana investigated. These transferable SSRs will facilitate future characterization of genomic diversity, relatedness and evolution across the entire Nicotiana genus.
The great value of SSR markers is their co-dominance and allelic multiplicity. The information for predicted allele number and size were determined and provided for each NIX marker in each of the seven genomes, so this will be an immediately useful resource for marker-assisted selection and genetic studies. This is the first report of comparative marker amplification information across multiple genomes of the same genus, and our TMPD database makes it easy to search for and compare the information for each marker, which will facilitate the efficiency of NIX markers use to follow linked agronomic or other genetic traits in future studies. In total, ~ 94% of NIX markers amplify 1–2 alleles per genome. The remaining NIX markers may be less useful for application due to multiple copies in a single genome, which is a possible outcome of their presence in repeated sequences such as transposable elements. In the past decade, molecular breeding using DNA markers has become the norm with many crop species [40,41,42]. The informative NIX markers developed here can be effectively used in genotyping of populations, QTL identification of interesting traits, germplasm diversity analysis, and marker-assisted selection in Nicotiana.
A highly efficient experimental protocol for the use of SSR markers was developed here, as well. Because all NIX markers were designed for uniform PCR conditions, the PCR reaction can be conducted at the single annealing temperature of 60 °C. For the standard PCR setting, the user will just add DNA, primers, and water to our standard master mix. This will facilitate rapid PCR application. Our PCR-based experimental validation of 120 NIX markers across five species or varieties demonstrated that the method worked in 100% of the cases investigated. Thus, the PCR product scoring method for NIX is of high efficiency, high resolution and high throughput with little background noise. Unlike the broad/fuzzy peaks seen with much PCR/SSR analysis, the sharp peaks in our method may result from the fully functional Taq polymerase employed, which retains the 3′-5′ exonuclease activity. In addition, our PCR settings for NIX marker have a high stringency that leads to the exclusive production of specific PCR fragments.
Conclusions
We characterized SSR in seven genomes of Nicotiana genus, developed NIX markers for these genomes, and constructed an online database for using the information. The NIX markers greatly expand Nicotiana marker resources, thus providing an immediately useful tool for research and breeding. We demonstrated a novel protocol for SSR marker development and SSR utilization at the whole genome scale that can be applied to any lineage of organisms. We also developed the TMPD database to facilitate NIX marker utility.
Methods
Nicotiana genomic sequence resources
The genomic sequences of seven Nicotiana used here are from publicly available databases. The draft assembly (version 0.44) of the genome sequence for N. ben was retrieved from the web site of Solgenomics. The genome sequences of N. syl (assembly ID: GCA_000393655.1, NCBI Accession No. ASAF00000000), N. tom (assembly ID: GCA_000390325.1, Accession No. ASAG00000000), N. oto (Accession No. AWOL000000000.1), N. tab TN90 (Accession No. AYMY00000000.1), N. Tab K326 (Accession No. AWOJ00000000.1) and N. tab Basma Xanthi (Accession No. AWOK00000000.1) were retrieved from Genbank at NCBI using the web links as described previously [17].
SSR discovery and analysis
Perfect SSRs, consisting of perfect repetitions of the same kind of nucleotide unit, were mined from whole genomic sequences of Nicotiana species by using our published software package, GMATA [7] (http://sourceforge.net/projects/gmata/). The parameters of SSR unit length were set to at least 12 bp for mono-nucleotide repeats, and at least five tandem copies for 2 bp to 10 bp nucleotide motifs. The five tandem copies criterion was used from our previous experiments [43] and other recent publications [44, 45].
Primer and marker design
Primers were designed in the sequence regions flanking each SSR locus by using the GMATA software package [7]. These settings were: product size ranging from 120 to 400 bp, targeted Tm of 60 °C (57–62 °C), primer target length of 20 nt (18–25 nt), minimum GC of 40%, maximum GC of 65% and optimized GC at 50%, allowed maximum poly-X 4, maximum self-complementarity 6, maximum 3′ self-complementarity 2, and maximum Ns 1. The size range of PCR products was chosen to permit easy fragment scoring in gels and DNA fragment size analyzers such as the ABI3730X.
SSR markers were then computed from the potential primer pairs. All designed primers were investigated for sequence identity. If sequences of both forward and reverse primer were 100% identical to those of other primer pairs, the primer pair was considered as a redundant primer pair. Only unique primer pairs were kept. An SSR marker was then recorded after assigning a unique sequential marker ID with the NIX prefix for each unique primer pair.
In silico marker mapping to genomes
To map the designed marker to genomic sequences, the GMATA software package [7] was used to perform an in silico amplification by calling the e-PCR algorithm [46]. The e-PCR result was used to process the marker mapping information. The settings for e-PCR were margin 3000, no gap in primer sequence, no mismatch in primer sequence, allowed size range of 100–1000, word size (−w) 12 and contiguous word (−f) 1. The mapping information in the results includes mapped or unmapped markers, the alleles of each marker, and marker anchoring information in each sequence of a genome. All designed markers were used to map to sequences of each Nicotiana genome, in silico, separately.
Experimental PCR validation of SSR markers
To test whether the newly-designed SSR markers were amplifiable, experimental PCR was conducted in several Nicotiana species and varieties. Genomic DNA was extracted from leaves of N. ben, N. syl, N. tom, N. tab variety K326, and N. tab variety HD or N. glutinosa by using the DNeasy Plant Mini Kit (Qiagen, Cat. No. 69104). DNA was diluted to 30–50 ng/μl in water before use. 120 markers were randomly chosen from the predicted commonly amplifiable makers across seven Nicotiana genomes for validation by using PCR amplification (Additional file 3: Table S2). For 96 markers scored in agarose gel only, PCR was performed in 20 μl reaction volumes containing 30–50 ng (1 μl) DNA, 0.53 μM tailed forward primer, 0.53 μM reverse primer, and 10 μl Phusion Hot Start Flex 2X Master Mix (NEB, Cat. No. M0536S). For 24 markers used in further fragment scoring, a tailed forward primer was synthesized with tail sequence CGTTGTAAAACGACGGCCAGT added to the 5′ end of each marker’s forward primer. PCR was performed in 20 μl volumes containing 30–50 ng (1 μl) DNA, 0.13 μM tailed forward primer, 0.27 μM 5’ FAM or 5’ HEX florescent labeled primer CGTTGTAAAACGACGGCCAGT, 0.53 μM reverse primer, and 10 μl Phusion Hot Start Flex 2X Master Mix (NEB, Cat. No. M0536S). PCR amplification and amplicon scoring were conducted according to our previous description for UGSW markers [43]. 15 μl of remaining amplification product were resolved in a 2.5% agarose gel. The PCR product from 24 markers with tail were further scored in ABI3730X sequence for high resolution fragment screening.
Comparison between reported markers and NIX markers
All publicly available Nicotiana markers were downloaded from the websites. These data included all amplifiable N. tab SSR markers reported previously [27, 33], and a mixture of 489 and 308 SSR and COSII markers from N. acuminata and N. tom genetic maps, respectively [28]. Redundant markers were sought and removed based on sequence identity of both primers for all markers after merging into one data set. From this analysis, we obtained 5397 SSR markers and 387 SSR or COSII markers from previous studies. A BLASTN search against reported non-redundant marker sequences was conducted to investigate the novelty of NIX markers with BLASTN algorithm in BLAST+ software and the settings are at -m8 –b5 -v5 –e 1e-5 and at least 95% identity [47]. NIX markers were identified as redundant markers if one or more primer sequence matched with those of reported markers.
Abbreviations
- BX:
-
Basma Xanthi
- N. ben :
-
N. benthamiana
- N. oto :
-
N. otophora
- N. syl :
-
N. sylvestris
- N. Tab :
-
N. tabacum
- N. tom :
-
N. tomentosiformis
- NIX:
-
Nicotiana multiple (X) genome markers
- SSR:
-
Simple sequence repeat
- TMPD:
-
Tobacco markers & primers database
References
Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5(6):435–45.
Klintschar M, Dauber E-M, Ricci U, Cerri N, Immel U-D, Kleiber M, Mayr WR. Haplotype studies support slippage as the mechanism of germline mutations in short tandem repeats. Electrophoresis. 2004;25(20):3344–8.
Forster P, Hohoff C, Dunkelmann B, Schürenkamp M, Pfeiffer H, Neuhuber F, Brinkmann B. Elevated germline mutation rate in teenage fathers. Proc R Soc Lond [Biol]. 1803;282:2015.
Kumar M, Choi JY, Kumari N, Pareek A, Kim SR. Molecular breeding in Brassica for salt tolerance: importance of microsatellite (SSR) markers for molecular breeding in Brassica. Front Plant Sci. 2015;6:688.
Shi J, Huang S, Fu D, Yu J, Wang X, Hua W, Liu S, Liu G, Wang H. Evolutionary dynamics of microsatellite distribution in plants: insight from the comparison of sequenced Brassica, Arabidopsis and other angiosperm species. PLoS One. 2013;8(3):e59988.
Zhao Z, Guo C, Sutharzan S, Li P, Echt CS, Zhang J, Liang C. Genome-wide analysis of tandem repeats in plants and green algae. G3. 2014;4(1):67–78.
Wang X, Wang L. GMATA: an integrated software package for genome-scale SSR mining, marker development and viewing. Front Plant Sci. 2016;7:1350.
Zhang Z, Deng Y, Tan J, Hu S, Yu J, Xue Q. A genome-wide microsatellite polymorphism database for the Indica and Japonica rice. DNA Res. 2007;14(1):37–45.
Song Q, Jia G, Zhu Y, Grant D, Nelson RT, Hwang E-Y, Hyten DL, Cregan PB. Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci. 2010;50(5):1950–60.
Sonah H, Deshmukh RK, Sharma A, Singh VP, Gupta DK, Gacche RN, Rana JC, Singh NK, Sharma TR. Genome-wide distribution and organization of microsatellites in plants: an insight into marker development in Brachypodium. PLoS One. 2011;6(6):e21298.
Xu J, Liu L, Xu Y, Chen C, Rong T, Ali F, Zhou S, Wu F, Liu Y, Wang J, et al. Development and characterization of simple sequence repeat markers providing genome-wide coverage and high resolution in maize. DNA Res. 2013;20(5):497–509.
Pandey G, Misra G, Kumari K, Gupta S, Parida SK, Chattopadhyay D, Prasad M. Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet Setaria italica (L.). DNA Res. 2013;20(2):197–207.
Shi J, Huang S, Zhan J, Yu J, Wang X, Hua W, Liu S, Liu G, Wang H. Genome-wide microsatellite characterization and marker development in the sequenced Brassica crop species. DNA Res. 2014;21(1):53–68.
Wang Q, Fang L, Chen J, Hu Y, Si Z, Wang S, Chang L, Guo W, Zhang T. Genome-wide mining, characterization, and development of microsatellite markers in Gossypium species. Sci Rep. 2015;5:10638.
Ow DW, De Wet JR, Helinski DR, Howell SH, Wood KV, Deluca M. Transient and stable expression of the firefly Luciferase gene in plant cells and transgenic plants. Science. 1986;234(4778):856–9.
Senthil-Kumar M, Mysore KS. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011;16(12):656–65.
Wang X, Bennetzen JL. Current status and prospects for the study of Nicotiana genomics, genetics, and nicotine biosynthesis genes. Mol Gen Genomics. 2015;290(1):11.
Sierro N, Battey J, Ouadi S, Bovet L, Goepfert S, Bakaher N, Peitsch M, Ivanov N. Reference genomes and transcriptomes of Nicotiana sylvestris and Nicotiana tomentosiformis. Genome Biol. 2013;14(6):R60.
Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller L, Martin G. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant-Microbe Interact. 2012;25(12):1523–30.
Sierro N, Battey JND, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV. The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun. 2014;5:3833.
Sierro N, van Oeveren J, van Eijk MJT, Martin F, Stormo KE, Peitsch MC, Ivanov NV. Whole genome profiling physical map and ancestral annotation of tobacco Hicks broadleaf. Plant J. 2013;75(5):880–9.
Renny-Byfield S, Chester M, Kovařík A, Le Comber SC, Grandbastien MA, Deloger M, Nichols RA, Macas J, Novák P, Chase MW, et al. Next generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol Biol Evol. 2011;28(10):2843–54.
Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann Bot. 2008;101(6):805–14.
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12(7):499–510.
Mauro-Herrera M, Wang X, Barbier H, Brutnell TP, Devos KM, Doust AN. Genetic control and comparative genomic analysis of flowering time in Setaria (Poaceae). G3. 2013;3(2):283–95.
Tong Z, Yang Z, Chen X, Jiao F, Li X, Wu X, Gao Y, Xiao B, Wu W. Large-scale development of microsatellite markers in Nicotiana tabacum and construction of a genetic map of flue-cured tobacco. Plant Breed. 2012;131(5):674–80.
Bindler G, Plieske J, Bakaher N, Gunduz I, Ivanov N, Van der Hoeven R, Ganal M, Donini P. A high density genetic map of tobacco (Nicotiana tabacum L.) obtained from large scale microsatellite marker development. Theor Appl Genet. 2011;123(2):219–30.
Wu F, Eannetta N, Xu Y, Plieske J, Ganal M, Pozzi C, Bakaher N, Tanksley S. COSII genetic maps of two diploid Nicotiana species provide a detailed picture of synteny with tomato and insights into chromosome evolution in tetraploid N. tabacum. Theor Appl Genet. 2010;120(4):809–27.
Moon HS, Nicholson JS, Lewis RS. Use of transferable Nicotiana tabacum L. microsatellite markers for investigating genetic diversity in the genus Nicotiana. Genome. 2008;51(8):547–59.
Morgante M, Hanafey M, Powell W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet. 2002;30(2):194–200.
SanMiguel P, Tikhonov A, Jin Y-K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, et al. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274(5288):765–8.
Temnykh S, DeClerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11(8):1441–52.
Bindler G, van der Hoeven R, Gunduz I, Plieske J, Ganal M, Rossi L, Gadani F, Donini P. A microsatellite marker based linkage map of tobacco. Theor Appl Genet. 2007;114(2):341–9.
Wu F, Eannetta N, Xu Y, Durrett R, Mazourek M, Jahn M, Tanksley S. A COSII genetic map of the pepper genome provides a detailed picture of synteny with tomato and new insights into recent chromosome evolution in the genus Capsicum. Theor Appl Genet. 2009;118(7):1279–93.
Zhang S, Tang C, Zhao Q, Li J, Yang L, Qie L, Fan X, Li L, Zhang N, Zhao M, et al. Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in foxtail millet Setaria italica (L.) P. Beauv. BMC Genomics. 2014;15:78.
Kumari K, Muthamilarasan M, Misra G, Gupta S, Subramanian A, Parida SK, Chattopadhyay D, Prasad M. Development of eSSR-markers in Setaria italica and their applicability in studying genetic diversity, cross-transferability and comparative mapping in millet and non-millet species. PLoS One. 2013;8(6):e67742.
Vasquez A, Lopez C. In silico genome comparison and distribution analysis of simple sequences repeats in cassava. Int J Genomics. 2014;2014:471461.
Yu J, Dossa K, Wang L, Zhang Y, Wei X, Liao B, Zhang X. PMDBase: a database for studying microsatellite DNA and marker development in plants. Nucleic Acids Res. 2017;45(D1):D1046–53.
Tong Z, Jiao T, Wang F, Li M, Leng X, Gao Y, Li Y, Xiao B, Wu W. Mapping of quantitative trait loci conferring resistance to brown spot in flue-cured tobacco (Nicotiana tabacum L.). Plant Breed. 2012;131(2):335–9.
Goutam U, Kukreja S, Yadav R, Salaria N, Thakur K, Goyal AK. Recent trends and perspectives of molecular markers against fungal diseases in wheat. Front Microbiol. 2015;6:861.
Miah G, Rafii M, Ismail M, Puteh A, Rahim H, Islam K, Latif M. A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci. 2013;14(11):22499.
Mir R, Zaman-Allah M, Sreenivasulu N, Trethowan R, Varshney R. Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor Appl Genet. 2012;125(4):625–45.
Serba D, Wu L, Daverdin G, Bahri B, Wang X, Kilian A, Bouton J, Brummer EC, Saha M, Devos K. Linkage maps of lowland and upland tetraploid switchgrass ecotypes. Bioenerg Res. 2013;6(3):953–65.
Yu J, Dossa K, Wang L, Zhang Y, Wei X, Liao B, Zhang X. PMDBase: a database for studying microsatellite DNA and marker development in plants. Nucleic Acids Res. 2016;45(D1):D1046–D1053. https://doi.org/10.1093/nar/gkw906.
Beier S, Thiel T, Munch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33(16):2583–85.
Schuler GD. Sequence mapping by electronic PCR. Genome Res. 1997;7(5):541–50.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinf. 2009;10:421.
Funding
This research was supported by funds from the Kunming Institute of Botany, Chinese Academy of Sciences (CAS) and the Foundation of Shandong Province Modern Agricultural Technology System Innovation Team from the Shandong Agricultural University (No. SDAIT-25-02) . The funding body did not have a role in the design of the study, data collection, analysis, interpretation of data, writing the manuscript, nor the decision to publish.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article, its Additional files 1, 2, 3 and 4, or The Tobacco Markers & Primers Database (TMPD) available at http://biodb.sdau.edu.cn/tmpd/index.html.
Author information
Authors and Affiliations
Contributions
XW, JLB and LY designed the study and wrote the manuscript. XW, SZ and ML conducted computing analysis. SY, XW and LY constructed the database. XW, YC, QZ, and YG conducted the experimental validation. XW and JLB had primary responsibility for revising the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The plants used in the research experiments were grown in Yunnan Academy of Agricultural Sciences according to the institutional and national guidelines.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1 Genomic SSR distribution of Nicotiana. (XLS 469 kb)
Additional file 2:
Figure S1 SSR length distribution. (PDF 364 kb)
Additional file 3:
Table S2 Duplication between NIX and reported markers. (XLS 41 kb)
Additional file 4:
Table S3 Information for experimentally validated markers. (XLS 51 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wang, X., Yang, S., Chen, Y. et al. Comparative genome-wide characterization leading to simple sequence repeat marker development for Nicotiana. BMC Genomics 19, 500 (2018). https://doi.org/10.1186/s12864-018-4878-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-4878-4