- Research article
- Open access
- Published:
Estrogen receptor beta impacts hormone-induced alternative mRNA splicing in breast cancer cells
BMC Genomics volume 16, Article number: 367 (2015)
Abstract
Background
Estrogens play an important role in breast cancer (BC) development and progression; when the two isoforms of the estrogen receptor (ERα and ERβ) are co-expressed each of them mediate specific effects of these hormones in BC cells. ERβ has been suggested to exert an antagonist role toward the oncogenic activities of ERα, and for this reason it is considered an oncosuppressor. As clinical evidence regarding a prognostic role for this receptor subtype in hormone-responsive BC is still limited and conflicting, more knowledge is required on the biological functions of ERβ in cancer cells. We have previously described the ERβ and ERα interactomes from BC cells, identifying specific and distinct patterns of protein interactions for the two receptors. In particular, we identified factors involved in mRNA splicing and maturation as important components of both ERα and ERβ pathways. Guided by these findings, here we performed RNA sequencing to investigate in depth the differences in the early transcriptional events and RNA splicing patterns induced by estradiol in cells expressing ERα alone or ERα and ERβ.
Results
Exon skipping was the most abundant splicing event in the post-transcriptional regulation by estradiol. We identified several splicing events induced by ERα alone and by ERα + ERβ, demonstrating for the first time that ERβ significantly affects estrogen-induced splicing in BC cells, as revealed by modification of a subset of ERα-dependent splicing by ERβ, as well as by the presence of splicing isoforms only in ERβ + cells. In particular, we observed that ERβ + BC cell lines exhibited around 2-fold more splicing events than the ERβ- cells. Interestingly, we identified putative direct targets of ERβ-mediated alternative splicing by correlating the genomic locations of ERβ and ERα binding sites with estradiol-induced differential splicing in the corresponding genes.
Conclusions
Taken together, these results demonstrate that ERβ significantly affects estrogen-induced early transcription and mRNA splicing in hormone-responsive BC cells, providing novel information on the biological role of ERβ in these tumors.
Background
Breast cancer (BC) is the most frequent cancer in women worldwide [1], and its development and progression are known to rely strongly on the stimulation by female sexual hormones, especially estrogens. Estrogen receptors α (ERα) and β (ERβ) are transcription factors that mediate the actions of estrogens in target cells [2,3]. Ligand binding to ERα or ERβ induces receptor dimerization, either as homodimers (ERα/ERα or ERβ/ERβ) or heterodimers (ERα/ERβ) [4], and promotes its translocation to the nucleus and binding of target chromatin sites via Estrogen Response Elements (EREs) and other regulatory elements on DNA [5]. Although encoded by different genes, ERα and ERβ share the same general modular protein structure of the nuclear hormone receptor superfamily, and have almost 100% amino acid sequence homology in their DNA-binding domain. They also show 59% amino acid homology in their ligand-binding domains [4,5]. The two ERs are thus quite similar in sequence and structure, but ERβ has considerably different and, in most cases, opposite biological effects compared to ERα in BC cells, both in vivo and in vitro. ERα regulates the transcription of hundreds of genes [6], enhancing BC cell growth, proliferation and survival in response to estrogens [7]. The specific role of ERβ and its impact in BC are unclear. This ER subtype is expressed in 70% of human breast tumors in combination with ERα, even if some human breast tumors express only ERβ [8-10]. Several reports have suggested that ERβ has anti-proliferative action in BC cells, by increasing the expression of anti-proliferative genes and/or decreasing the expression of proliferative and anti-apoptotic genes [11-15] and the ERα/ERβ ratio determines the cell-specific response to estrogen. In BC this ratio is higher than in normal tissues, due to up-regulation of ERα and down-regulation of ERβ [16]. Loss of ERβ mRNA levels in cancer can occur as a result of promoter methylation [17]. These observations have suggested a positive prognostic value of this receptor subtype [12,18]. However, several studies have reported also a negative prognostic value for ERβ expression [19,20], making the overall contribution of this receptor isoform to BC biology unclear.
The exact mechanism of the antagonism between ERβ and ERα is only partially known. As the two receptors share only 30% homology in their transactivation domain AF1 [5], it is likely that they show different patterns of interaction with coregulatory proteins. Indeed, we have previously reported that the protein interactomes of both ERβ and ERα show significant differences of the protein complexes engaged by the two ER subtypes [21-25]. A particularly interesting subset of interacting proteins, with only partially overlapping interaction patterns between the two receptors [26], comprises factors involved in RNA maturation and splicing [Additional file 1: Figure S1] [26].
Alternative splicing is a mechanism by which cells can increase the variability of their proteomes by changing the composition of transcribed genes through differential choice of exons to be included in the final mRNA molecule [27]. Almost 90% of human genes show alternative splicing during development, cell differentiation and disease [28]. Recent studies have shown the existence of cancer-specific splicing events by which transformed cells switch from the adult isoform of the gene to a more embryonic one, contributing to the cancer phenotype [29-31].
Alternative splicing events have been monitored in BC and in numerous tumor types [32], and ERα itself has been reported to induce alternative splicing of a specific set of genes [33-35]. Here, we investigated the ability of ERβ to regulate mRNA maturation and splicing in hormone-responsive BC cells. To this purpose, we performed high-throughput RNA sequencing (RNA-seq) analysis of human MCF-7 cell lines stably transfected with ERβ, and compared them with the wild type line, expressing only ERα, upon 17β-estradiol (E2) stimulation.
Results
High-throughput sequencing in ERβ + and ERβ- human BC cell lines
We have previously established and characterized subclones of the human BC cell line MCF-7 expressing human ERβ fused to a Tandem Affinity Purification (TAP) tag, and have shown that the addition of the TAP-tag at either the N- or the C-terminus of the protein (indicated as Nt-ERβ and Ct-ERβ, respectively) does not alter significantly the receptor function, nor its ability to activate transcription or to antagonize ERα-dependent transcription [21,22,36]. In order to study the early events of hormone-induced pre-mRNA maturation, we used these stable cell lines to perform deep-sequencing analysis of estrogen-induced transcriptional events shortly after stimulation with 17β-estradiol (2 h), to focus mostly on primary transcriptional events [21,36]. For comparison, we also performed the experiment in wild-type MCF-7 cells, which do not express endogenous ERβ.
Almost 70 million reads/replicate were aligned against the reference human genome for ERβ- and ERβ + BC cell lines. The number of reads for genes and isoforms were normalized to “Fragment Per Kilobases of exon per Million of mapped reads” (FPKM). In order to analyze genes and isoforms, we set 0.5 FPKM (at least one analyzed condition, with/without E2 stimulus) as the expression level threshold. In this way, we identified 16,821 (MCF-7 wt), 16,148 (Ct-ERβ) and 17,135 (Nt-ERβ) genes as expressed. The criteria for considering genes and isoforms as significantly regulated by estradiol were: FPKM value ≥0.5 in at least one analyzed condition, q-value (FDR-adjusted p-value of the test statistics) ≤0.05 and |fold-change| (FC) ≥1.3 [Additional file 2: Table S1]. As shown in Figure 1, 895 (MCF-7 wt), 2,899 (Ct-ERβ) and 3,043 (Nt-ERβ) genes were detected as significantly regulated by E2 in these BC cell lines. Expression of ERβ in MCF7 cells significantly affected the estrogen-dependent gene expression profile: the regulation of around 230 genes (≈25% of E2-regulated genes) was lost in both cell lines expressing ERβ, while a large number of genes which were not regulated in wt cells became significantly regulated in Ct-ERβ (2,396) and Nt-ERβ (2,463) clones (Figure 1). The genes regulated consistently in both ERβ + lines are reported in Additional file 2: Tables S1-D. Interestingly, expression of ERβ in this BC cell line had a stronger effect on inhibited genes than on the activated ones: regulation of 40% of genes inhibited by estradiol in wt cells was lost in both ERβ + cell lines, versus 14% for estrogen-activated genes. Gene Ontology analysis revealed that among the most enriched functions in the group of genes whose regulation by estradiol was lost in both ERβ + cells there were, as expected, DNA Replication, Recombination and Repair, as well as Cell Cycle and Cell Morphology (data not shown).
Venn diagram of expressed genes in ERβ + and wt breast cancer cell lines. The Venn diagram shows the number of genes regulated by estradiol in the three cell lines, as indicated: wt (parental MCF-7 cells); Ct-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the C-terminus); Nt-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the N-terminus). The pie charts in the lower panels specify the direction of regulation by estradiol (induction or repression) of the genes whose regulation is present in wt cells but is lost in both ERβ + cell lines (left panel), or of the genes whose regulation by estradiol is not present in the wt cells but appears in both ERβ + lines (right panel).
Estrogen-dependent splicing events in ERβ + and ERβ- human BC cell lines
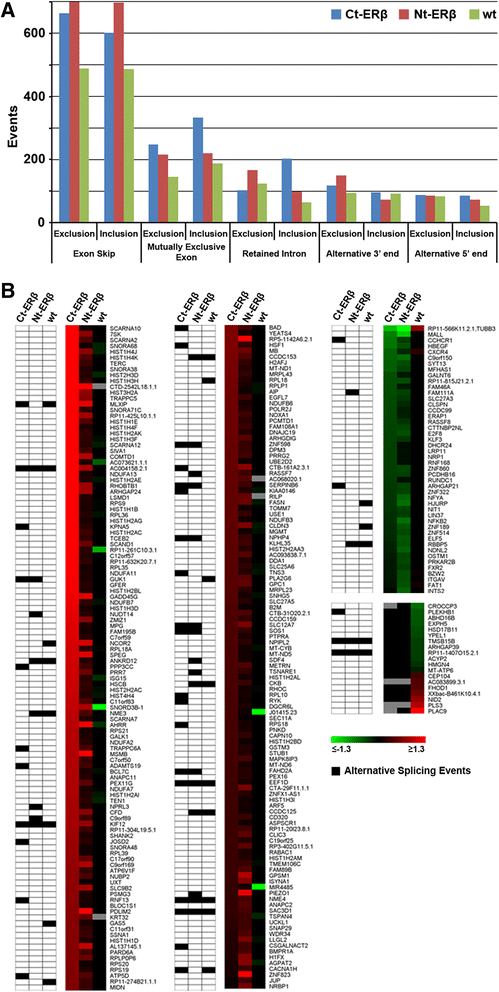
Events of exon skipping, mutually exclusive exons, alternative start, stop splice site and intron retention were annotated using the Multivariate Analysis of Transcript Splicing (MATS) software [37] [Additional file 3: Tables S2]. Exon skipping appeared to be the predominant splice event in all cell lines analyzed. MATS reveled in detail 1,264 (Ct-ERβ), 1,402 (Nt-ERβ) and 975 (MCF-7 wt) exon skipping events induced by estradiol, associated with 1,016 (Ct-ERβ), 1,117 (Nt-ERβ) and 816 (wt MCF-7) genes (Figure 2A). Five hundred seventy-five events were common to all the cell lines analyzed while 115 showed opposite exon inclusion level in ERβ + lines compared to ERβ- wt cells. We also observed high levels of retained intron and mutually exclusive exons events, confirming a complex and significant effect of ERs on the regulation of RNA splicing in these cell lines (Figure 2A).
Annotation of splice events in ERα + and ERα + ERβ + BC cell lines. (A) The bar plot shows the number of all alternative splicing events occurring in the cell lines analyzed. Inclusion and exclusion behavior for each event are shown (FDR ≤ 0.05; c ≤ |0.1|). (B) Genes whose regulation has opposite direction in the ERβ + lines compared to the wt MCF-7. The heat map on the right side shows the gene expression fold changes induced by estradiol. The matrix on the left side shows in black those genes for which a splicing event was detected in at least one of the cell lines. The nomenclature for the cell lines is the following: wt (parental MCF-7 cells); Ct-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the C-terminus); Nt-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the N-terminus).
To focus on the differences in splicing patterns between ERβ + and ERβ- cell lines, we first looked at the genes which were regulated by estradiol in opposite direction in ERβ + cells versus wt cells [Additional file 4: Table S3]. Among 298 regulated genes, 56 also underwent estradiol-induced alternative splicing in at least one of the cell lines, confirming that pre-mRNA maturation was regulated concurrently with transcription in a significant fraction of ERβ-regulated genes (Figure 2B). These ERβ-regulated genes undergoing alternative splicing included transcriptional regulators (NCOR2, ZNF189, MLXIP, ANKRD12, HSF1), enzymes involved in nucleoside/nucleotide metabolism (GUK1, NME3, NME4), actin remodeling and cellular transport processes (TNS3, TRAPPC6A, TMSB15B, KIF12), and protein translation (ZNF98, EEF1D, RPL10, RPL18, RPS18).
Estrogen-induced differential splicing has been reported also in genes independently on transcriptional regulation [35]. In order to find the splicing events differentially regulated by estradiol in ERβ + compared to wt cells, we scanned the whole list of expressed genes for splicing patterns occurring differentially in ERβ + vs ERβ- cells. In addition, we focused on those splicing events whose occurrence significantly altered the ratio between different isoforms of the same gene. Therefore, for each isoform we calculated the percentage of gene expression associated with that particular isoform (FPKM ratio: FPKMisoform/FPKMgene %) and selected for further analysis only those isoforms in which estradiol induced a change in FPKM ratio of at least 10% either in wt MCF-7 or in both ERβ + cell lines. Other criteria of inclusion were: occurrence of at least one splicing event as detected by MATS; at least one isoform of the gene with FPKM value ≥0.5 in at least one analyzed condition, q-value ≤0.05 either in wt or in both ERβ + cell lines; regulation in both ERβ + lines in opposite directions compared to the wt. In this way, we identified the quantitatively most relevant splicing events differentially regulated by E2 in ERβ + versus ERβ- cells, including 35 genes whose isoform composition changed significantly after E2 stimulation in an ERβ-dependent fashion (Figure 3). Among these, we found genes involved in apoptosis (BAD), lipid metabolism (ACADM, PLSCR1, SLC27A2, STARD4), nutrient transport (SLC25A19, SLC35C2), transmembrane receptor signaling (IFNGR2, LDLRAD4), Notch signaling (PSEN2, POGLUT1, SGK1, SLC35C2), as well as some non-coding RNAs (MCM3AP-AS1, SNHG17). An example of a gene whose splicing pattern was affected by estradiol in an ERβ-dependent fashion is reported in Figure 4A, showing the gene SGK1, which encodes a serum and glucocorticoid-induced serine/threonine protein kinase involved in ion transport affecting many cellular processes such as cell growth, proliferation, survival, apoptosis and migration [38,39]. The gene was induced by estradiol in both wt and ERβ + cells; however, expression of ERβ in the absence of hormonal stimulation induced a promoter usage switch and retention of the first intron causing an alternative translation start site and therefore the expression of an isoform with a different N-terminal sequence (ENST00000367857), compared to the major isoform expressed (ENST00000237305). It has been reported that alternative isoforms at the N-terminus can affect SGK1 localization and protein stability: the ERβ-specific form with intron-retention misses a very crucial sequence involved in targeting to the endoplasmic reticulum as well as in proteasomal degradation [40].
Selected isoform switches affected by the expression of ERβ. The ERβ-dependent differential splicing events that affect most prominently the balance of different isoforms for each gene were identified by using the following parameters: (i) isoform ratio (FPKMisoform/FPKMgene %) changing of at least 10% after estradiol stimulation, either in the wt cells or in both the ERβ + cells in at least one of the isoforms of the gene; (ii) at least one isoform of the gene significantly regulated by estradiol (p-value ≤ 0.05; |FC| ≥ 1.3) either in the wt cells or in both the ERβ + cells; (iii) isoform ratio changing in opposite direction in the wt cells compared to the ERβ + cells; (iv) at least one splicing event identified by MATS analysis. Thirty-five genes satisfied all the selection requirements; of these, only 2 to 3 isoforms were selected for presentation, according to the expression levels and the regulation by estradiol. Left panel: heat map of regulation of the selected genes by estradiol in: Ct-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the C-terminus); Nt-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the N-terminus); wt (parental MCF-7 cells). Right panel: heat map with the change in isoform ratio (FPKMisoform/FPKMgene %) between E2-treated and non-treated cells in the indicated cell lines; for each gene, at least two different isoforms are presented, to show estrogen-induced switch from one isoform to the other.
Examples of ERβ-specific splicing events. A) Example of alternative splicing in the SGK1 gene. The upper panel shows a schematic representation of two differentially regulated isoforms of the gene (same as those shown in Figure 3 for the same gene), differing in the transcription start site, in the inclusion of the first intron (the gene is encoded by the reverse strand) and in the transcription stop site. The lower panels show a representation of the RNA-Seq reads and junction reads associated with the gene in the different conditions: Ct-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the C-terminus) without or with E2 stimulation, and wt (parental MCF-7 line) without or with E2 stimulation. B) Example of ERβ-specific alternative promoter usage in the gene PSD3. The left panel shows a heat map with the fold change of all the primary transcripts associated with the gene in: Ct-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the C-terminus); Nt-ERβ (MCF-7 subclone stably expressing ERβ tagged with the TAP-tag at the N-terminus); wt (parental MCF-7 cells). The right panels show the logarithm base 10 (log10) FPKM for each of the different transcript start sites (from TSS01 to TSS09) corresponding with the different gene isoforms in the above described cell lines, without (left) or with (right) E2 stimulation.
This data suggests that expression of ERβ causes switches in estradiol-induced splicing patterns, potentially affecting expression or function or ER targets.
Estrogen-dependent alternative promoter usage in ERβ + vs ERβ- BC cells
As the usage of alternative promoters is a major determinant of protein diversity, even more than alternative splicing [41,42], we next focused on utilization of multiple promoters. We grouped the primary transcripts of a gene based on the promoter used, and subsequently tested changes in primary transcript abundance by measuring the square root of the Jensen-Shannon divergence that occurred within and between the analyzed groups. Finally, we investigated the potential promoter switch regulation for the genes comprising more than one differentially expressed transcript initiating from distinct genomic loci. There were 977 (Ct-ERβ), 402 (Nt-ERβ) and 222 (wt MCF-7) distinct promoter-switching genes (FDR ≤ 0.05) in ERβ + and ERβ- BC cell lines, respectively [Additional file 5: Tables S4A-C]. Of the 222 promoter-switching genes recorded in wt MCF-7 cells, 165 did not show promoter switch in both ERβ + cell lines, while 61 new promoter-switching events, not present in wt cells, were detected in both ERβ + lines. These 61 ERβ-specific promoter-switching genes are involved in important cellular functions known to be controlled by E2 in BC cells, such as transcription (FOXJ3, GTF2H, NR2C2AP), DNA metabolism and repair (PRKDC, REV3L, SCAND3), pre-mRNA maturation and splicing (PPM1G, PRPF38B, RNMT, RPRD1A, SMG1), translation (FARSB, RARS, RPS21, UTP20), protein ubiquitination and proteasome pathway (CUL5, KLHL2, PSMB1, USP7), cytoskeleton and cytokinesis (DCTN4, MYL12A, SEPT9, SYDE2), membrane metabolism, remodeling and intracellular transport (ATP9A, CAST, MAL2, PSD3, TMEM43, TRAPPC9), cell adhesion and polarity (ARHGAP12, CD9, CLDN7, EPB41L5, PERP), signal transduction (MAP3K5, NGFRAP1, TNFRSF12A, WWC3, ZDHHC5).
As an example, we focused on the PSD3 gene, predicted to be a nucleotide exchange factor for ADP Ribosylation Factor (ARF) 6, a member of the RAS family involved in vesicular trafficking, remodeling of membrane lipids, and signal transduction [43]. As show in Figure 4B, for this gene we found 9 distinct primary transcripts (TSS01-TSS09) whose usage ratios changed with E2 stimulus (right panel) compared to the control untreated cells (left panel). In ERβ + cells, estradiol induced a switch from promoter TSS08 (ENST00000523619) to the downstream promoter TSS02, resulting in a shorter transcript (ENST00000519653), which is predicted to undergo nonsense-mediated decay [Additional file 6: Table S5].
Estrogen-dependent splice ratios in ERβ + vs ERβ- cells
To obtain a comprehensive view of the estrogen-induced differences in the splice ratios between wt, Ct-ERβ and Nt-ERβ cells, we employed Cuffdiff v2.1.1 [44], which calculates the changes in splice isoforms abundance, by quantifying the square root of the Jensen-Shannon divergence, considering each primary transcripts able to produce multiple isoforms. We determined 217 (Ct-ERβ), 241 (Nt-ERβ) and 95 (MCF-7 wt) differentially spliced genes (DSGs, i.e. genes for which estradiol challenge induced at least one splicing event) with a FDR value <0.05 [Additional file 5: Tables S4D-F]. Of the 95 spliced genes detected in wt cells, sixty-nine lacked splicing events in both ERβ + cell lines, suggesting inhibition of ERα-dependent splicing by ERβ. Moreover, 28 ERβ-specific DSGs, in which no estrogen-induced splicing was recorded in wt cells, were found in both ERβ + cell lines. These include genes involved in mitosis and cytokinesis (SEPT9, BICD2, ENSA, PDS5A), cell cycle control (CCNJ, RAN), transcription (C11orf30, EIF3M, HIPK1, PBX1, ZNF124, ZNF131), protein folding (DNAJB6, HSP90B1), ubiquitination and sumoylation (DCUN1D4, SENP5, TRIM33), and signal transduction (APBB2, RTKN2).
Correlation between ER binding and ER-dependent splicing
In order to identify direct splicing targets of ERα and ERβ, we next investigated the presence of ERα and ERβ binding sites in genomic locations close to the identified DSGs [Additional file 7: Table S6]. To verify the specificity of ER binding sites, we created a heat-map based on previously published ChIP-Seq data [21,45]. The binding densities of ERα and ERβ were clustered according to the seqMINER platform [46]. In the clustering shown in [Additional file 8: Figure S2], each line represents a genomic location of a binding site with its surrounding ±1.5 kb region. In the left panel, ERα binding sites were used as a reference to collect a ChIP-seq tag densities window in ERβ + and ERβ- cell lines, while in the right panel ERβ binding sites were used as a reference. The heat map, representing the clustered density matrix, confirmed that ERβ presence modified a significant number of ERα binding sites. In order to investigate the correlation between ERα and ERβ DNA binding and splicing events, we compared our RNA-Seq data with the ChIP-Seq data we had previously obtained in these same cell lines [21]. We considered the binding sites included within regions spanning 10 kb upstream or downstream of all DSGs in each cell line. The Circos plot [47] in Figure 5 shows the DSGs for which estradiol challenge induced at least one splicing event in both ERβ + cell lines with a nearby binding site for ERα and/or ERβ. The outer ring (blue) reports the ERα binding sites, while the inner ring (red) shows the ERβ binding sites. Based on ERα and ERβ binding, we distinguished three different DSGs groups. Group 1 DSGs were associated with both ERα and ERβ binding sites (black). Group 2 includes DSGs associated with ERα binding sites exclusively (blue) and group 3 DSGs associated with ERβ binding sites exclusively (red). The vast majority of genes exhibited both binding sites (Group 1), confirming the competing role of ERα and ERβ. Interestingly, among the putative direct ERβ splicing targets we found many genes involved in transcription: transcription factors (FOXN3, NFIB, TAF6, TCF12, ZNF295, ZNF438), histone methyltransferases (ASH1L, MLL5, SETD5, SETMAR) and acetyltransferase (YEATS2), other transcriptional regulators (AKNA, BANP, CHD3, MEIS2). Other interesting functions associated with these genes are: apoptosis (BCL2L13, CASP8, C1orf201), autophagy (AMBRA1, ATG12, ATG13, ATG16L2), splicing (HNRNPH3, MBL2), protein ubiquitination (CNOT4, FBXW11, RFWD2, WDR20) cytoskeleton and cytokinesis (AURKA, EPS8L2, EPB41L2, LARP4, NPHP4, PLEC, TLN2), primary cilium biogenesis [48] (BBIP1, IFT140, KIAA0586), intracellular trafficking/membrane trafficking (ASAP1, KIF13A, MCOLN1, RAB17, TBC1D1, VPS29), cell adhesion (ARMC8, CD151, ELMOD3, LLP, VMP1).
Correlation between ERβ-specific splicing and ER binding. Circos plot of differential spliced genes (DSGs) common to both ERβ + cell lines which contain at least one ER binding event within a window of 10 kB around the gene. The outer ring shows chromosome ideograms with the relative genes located in their respective chromosomal locations, with the following color code: the genes that have both ERβ and ERα binding are in black; the genes which have only ERβ binding are in red; the genes which have only ERα binding are in blue. The two internal rings represent ERα and ERβ binding events in blue and red, respectively.
Taken together, these analyses suggest the strong effect of ERs binding on alternative splicing and confirming the key role of ERβ in BC cells.
Discussion
In this study we investigated for the first time the effects of ERβ expression on estrogen-dependent pre-mRNA maturation and splicing. We used two different subclones of ERα-positive and estrogen-dependent MCF-7 cells stably expressing ERβ, and we found that expression of ERβ in these cells significantly affected the estradiol-dependent early transcriptional program and splicing pattern. In particular, introduction of ERβ caused loss of regulation of 25% of the estrogen-regulated genes. Moreover, a comparison between the ERβ + and the wt ERβ- cells showed that expression of ERβ caused loss of ERα-induced promoter switching in 75% of the genes, and of ERα-induced splicing in 72% of the genes.
Besides affecting ERα-dependent transcription and splicing, expression of ERβ caused the appearance of ERβ-specific estrogen-responsive transcription (550 genes) [Additional file 2: Tables S1-D], promoter-switching (61 genes) and differential splicing (28 genes) events, counting only the events that were present in both ERβ + lines. The biological functions of the ERβ-specific genes included DNA replication and repair, cell cycle, apoptosis and autophagy, DNA transcription, lipid metabolism, membrane metabolism, intracellular trafficking, mRNA maturation and translation, protein ubiquitination and sumoylation, cell signaling, confirming that a wide variety of cellular processes are affected by ERβ.
Based on our data, there are at least three different mechanisms by which ERβ can affect ERα-dependent transcription: (1) competition with ERα for binding to target gene promoters, in the form of competitive binding or heterodimerization, which can alter the recruitment of coregulators: the comparison between Chip-seq and RNA-seq data showed that the majority of the primary targets identified in this experiment were directly targeted by both ERα and ERβ (Figure S1), as expected also from data in the literature [49]; (2) gain of new binding sites that are not bound by ERα alone: a subset of binding sites appeared in the ERβ + cells but were not present in wt cells, again consistently with a previous report comparing ERα and ERβ genomic binding patterns in MCF-7 cells [49]; (3) secondary effects: expression of ERβ induced transcription and splicing of transcriptional regulators (most interestingly, the corepressor NCOR2, involved in gene repression by tamoxifen-bound estrogen receptor and by unliganded NRs such as the retinoic acid receptors) and of splicing factors, which can in turn affect estrogen-induced transcription and pre-mRNA maturation. For instance, ERβ induced promoter switching in the PPM1G gene, encoding a protein phosphatase responsible for dephosphorylation of pre-mRNA splicing factors [50]; the ERβ-specific alternate protein isoform from this gene (ENST00000350803) has an additional 17 amino acids in the N-terminal catalytic domain compared to the main isoform (ENST00000544412), which are likely to modulate the enzymatic function.
Even if the sole detection of splicing events does not give information on the biological consequences of ERβ effects on RNA splicing, it is tempting to speculate on the possible implications of ERβ-dependent splicing events on BC cells. Changes in splicing patterns can affect biological processes by many different mechanisms, including gain-of-function or functional switches, altered cellular localization, dominant negative effect, changes in protein/mRNA stability. For instance, expression of ERβ induced a promoter switch in the USP7 gene, resulting in a shorter transcript (ENST00000535863) compared to the major isoform expressed in wt cells (ENST00000381886). This ERβ-specific isoform is missing the N-terminal 84 amino acids, a region of the protein of critical importance for interaction with substrates. This suggests the possibility of a switch in substrate affinity induced by ERβ. This is particularly interesting as USP7 is a deubiquitinating enzyme, responsible for removing ubiquitin chains from both the tumor suppressor p53 and its negative regulator Mdm2 [51], which instead is an ubiquitin ligase inducing degradation of p53. As USP7 binds both p53 and Mdm2 with the same N-terminal domain [51], the overall effect of its enzymatic activity is highly dependent on the relative affinity for the two targets. The ERβ-induced switch may alter this equilibrium, thus modulating such a relevant aspect of cancer biology as p53 stability. In the case of IFNγ Receptor 2, ERα induced expression of the full length transcript (ENST00000381995), while ERβ favored a switch toward truncated forms (ENST00000545369, ENST00000405436) lacking the transmembrane domain, and therefore predicted to be secreted as soluble forms in the extracellular environment, with the potential of acting as dominant negative modulators of interferon signaling. In another example (the gene PSEN1, involved in intramembrane proteolysis and cleavage of the intracellular domain of transmembrane proteins such as amyloid precursor protein and Notch and therefore a potential therapeutic target in BC [52]), the ERβ-induced splicing switch did not alter the open reading frame but caused the expression of a longer mRNA (ENST00000366783) compared to the isoform that was favored by ERα (ENST00000422240), possibly affecting the rate of translation or stability of the mRNA.
Another way of finding hints to the biological consequences of ERβ-specific splicing events described here is the reconstruction of pathways whose genes are differentially spliced following ERβ introduction in the cells. For instance, we found many genes involved in the Notch signaling pathway differentially spliced by ERβ: the above-mentioned PSEN2 is the catalytic subunit of the γ-secretase complex, responsible for intramembrane proteolysis of transmembrane receptors including Notch, resulting in the release of Notch Intracellular Domain (NICD), which migrates to the nucleus and regulates transcription of target genes [53]. Also, NCOR2 is bound to the unliganded CBF-1 transcription factor, a primary effector of Notch signaling which acts as a repressor in unstimulated cells, but is converted to an activator (dismissing the corepressor NCOR2) after binding the NICD [54]. Furthermore, POGLUT1 has recently been shown to be an endoplasmic reticulum O-glycosyl-transferase responsible for glycosylation of Notch and required for its function [55], and SLC35C2 is an endoplasmic reticulum transporter responsible for accumulation of GDP-fucose, which is used for Notch fucosylation, required for full activation [56]. Finally, SGK1 has recently been shown to be a negative regulator of Notch signaling by inhibiting γ-secretase activity and promoting Notch degradation [57], and MAGI1 has been shown to recruit Notch ligand Dll1 to cadherin-based adherens junctions, stabilizing it on the cell surface [58]. It is worth noting that the predominant Notch receptor expressed in these cell lines was Notch2, which was induced by E2 in wt cells (to a level slightly below the chosen cut-off threshold, but highly statistically significant: FPKM without E2: 49.826; FPKM with E2: 61.583, FC 1.236, q-value 0.0005), while its basal expression was lowered and its up-regulation smoothened in the ERβ + cells.
A possible modulation of the Notch pathway by ERβ is especially interesting as Notch is a known regulator of breast development and maintenance of breast stem cells [59]; alterations in the Notch pathway have been involved in breast carcinogenesis, and in particular the Notch pathway has been implicated in the development of triple negative BC (TNBC), a particularly aggressive form of BC which does not express ERα, progesterone receptor (PR) or HER2, and which has shown resistance to all known therapies [60]. Targeting Notch signaling has been proposed in TNBC. As these cancers are ERα-negative, hormonal treatment is not currently used for these patients; however, ERβ could be expressed in up to 50% of TNBCs [10,18], and its expression in TNBC has been associated with better prognosis [18]. Therefore, ERβ may represent a potential new therapeutic target in TNBC. The interrelation between ERβ and Notch in the development and prognosis of triple-negative BC should be investigated further in future research.
Conclusions
In conclusion, whole-genome analysis of early transcription evens and mRNA processing associated with ERβ confirmed a relevant role for this receptor in modulating ERα-dependent transcription and splicing, but also identified novel, ERβ-specific transcription and splicing events, confirming a wide range of actions of ERβ in the biology of BC.
The data reported here confirm the complexity of estrogen action in BC cells and provide a comprehensive description of the effects of ERα and by ERβ on early transcription and splicing in hormone-responsive BC cells. More importantly, they provide a starting point to identify the events of ERβ-dependent splicing which are most significant for cancer biology.
Methods
Human hormone-responsive BC cells
Stable cell clones expressing either C-TAP-ERβ or N-TAP-ERβ (ERβ+) generated as previously described [23], and wild type (wt) MCF7 (ERβ-) cells were used for this study. All cell lines were maintained, propagated, hormone-starved for 5 days and analyzed for estrogen signaling as described earlier [21,45].
Illumina RNA sequencing library preparation
Total RNA was extracted from hormone-starved cell cultures (+Ethanol -E2) or after 2 hours of stimulation with 10-8 M 17β-estradiol (+E2), as described previously [21]. RNA concentration in each sample was determined with a NanoDrop-1000 spectrophotometer and the quality assessed with the Agilent 2100 Bioanalyzer and Agilent RNA 6000nano cartridges (Agilent Technologies). Indexed libraries were prepared from 1 μg/ea. of purified RNA with TruSeq Stranded total RNA Sample Prep Kit (Illumina) according to the manufacturer’s instructions. Libraries were sequenced (paired-end, 2×100 cycles) at a concentration of 8 pmol/L per lane on HiSeq1500 platform (Illumina) with a coverage of >70 million sequence reads/sample on average.
Read alignment and transcript assembly
TopHat v.2.0.10 [61] was used to align all reads including junction-spanning reads back to the human genome (Homo sapiens Ensembl GRCh37, hg19). To identify the differentially expressed and spliced genes and isoforms between ERβ- and ERβ + cell lines we used Cuffdiff v2.1.1 [44]. The parameters to define genes and isoforms as differentially expressed were the following: expression level threshold of 0.5 FPKM; q-value (FDR-adjusted p-value of the test statistic) ≤ 0.05 and |FC| ≥ 1.3. Moreover, to detect the ERβ- and ERβ + BC-specific splice events such as exon skip, exon inclusion, alternative splice sites and intron retention, we performed a direct comparison analysis using MATS v3.0.8 [37]. To filter events with at least 10% change in exon inclusion level we set the MATS cutoff c, representing the extent of splicing change one wishes to identify, to 0.1, and FDR ≤ 0.05 to filter the identified splice events.
Data access
RNA-Seq data have been deposited in the Gene Expression Omnibus genomics data public repository (http://www.ncbi.nlm.nih.gov/geo/) with Accession Number GSE64590.
Abbreviations
- ER:
-
Estrogen receptor
- Nt-ERβ:
-
N-TAP-estrogen receptor beta
- Ct-ERβ:
-
C-TAP-estrogen receptor beta
- E2:
-
17β-estradiol
- BC:
-
Breast cancer
- FPKM:
-
Fragment per kilobases of exon per million
- DSGs:
-
Differentially spliced genes
- TSSs:
-
Transcripts starting sites
- DPU:
-
Differential promoter usage
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359-86.
Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, et al. Mechanisms of estrogen action. Physiol Rev. 2001;81(4):1535–65.
Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116(3):561–70.
Cowley SM, Hoare S, Mosselman S, Parker MG. Estrogen receptors alpha and beta form heterodimers on DNA. J Biol Chem. 1997;272(32):19858–62.
Pace P, Taylor J, Suntharalingam S, Coombes RC, Ali S. Human estrogen receptor beta binds DNA in a manner similar to and dimerizes with estrogen receptor alpha. J Biol Chem. 1997;272(41):25832–8.
Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145(4):622–34.
Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144(10):4562–74.
Kurebayashi J, Otsuki T, Kunisue H, Tanaka K, Yamamoto S, Sonoo H. Expression levels of estrogen receptor-alpha, estrogen receptor-beta, coactivators, and corepressors in breast cancer. Clin Cancer Res. 2000;6(2):512–8.
Speirs V, Carder PJ, Lane S, Dodwell D, Lansdown MR, Hanby AM. Oestrogen receptor beta: what it means for patients with breast cancer. Lancet. 2004;5(3):174–81.
Skliris GP, Leygue E, Curtis-Snell L, Watson PH, Murphy LC. Expression of oestrogen receptor-beta in oestrogen receptor-alpha negative human breast tumours. Br J Cancer. 2006;95(5):616–26.
Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. ER beta inhibits proliferation and invasion of breast cancer cells. Endocrinology. 2001;142(9):4120–30.
Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. Estrogen receptor beta inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res. 2004;64(1):423–8.
Strom A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA. Estrogen receptor beta inhibits 17beta-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci U S A. 2004;101(6):1566–71.
Chang EC, Frasor J, Komm B, Katzenellenbogen BS. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology. 2006;147(10):4831–42.
Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2008;27(7):1019–32.
Roger P, Sahla ME, Makela S, Gustafsson JA, Baldet P, Rochefort H. Decreased expression of estrogen receptor beta protein in proliferative preinvasive mammary tumors. Cancer Res. 2001;61(6):2537–41.
Zhao C, Lam EW, Sunters A, Enmark E, De Bella MT, Coombes RC, et al. Expression of estrogen receptor beta isoforms in normal breast epithelial cells and breast cancer: regulation by methylation. Oncogene. 2003;22(48):7600–6.
Honma N, Horii R, Iwase T, Saji S, Younes M, Takubo K, et al. Clinical importance of estrogen receptor-beta evaluation in breast cancer patients treated with adjuvant tamoxifen therapy. J Clin Oncol. 2008;26(22):3727–34.
Guo L, Zhu Q, Yilamu D, Jakulin A, Liu S, Liang T. Expression and prognostic value of estrogen receptor beta in breast cancer patients. Int Journal Clin Exp Med. 2014;7(10):3730–6.
Chantzi NI, Tiniakos DG, Palaiologou M, Goutas N, Filippidis T, Vassilaros SD, et al. Estrogen receptor beta 2 is associated with poor prognosis in estrogen receptor alpha-negative breast carcinoma. J Cancer Res Clin Oncol. 2013;139(9):1489–98.
Grober OM, Mutarelli M, Giurato G, Ravo M, Cicatiello L, De Filippo MR, et al. Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation. BMC Genomics. 2011;12:36.
Paris O, Ferraro L, Grober OM, Ravo M, De Filippo MR, Giurato G, et al. Direct regulation of microRNA biogenesis and expression by estrogen receptor beta in hormone-responsive breast cancer. Oncogene. 2012;31(38):4196–206.
Nassa G, Tarallo R, Ambrosino C, Bamundo A, Ferraro L, Paris O, et al. A large set of estrogen receptor beta-interacting proteins identified by tandem affinity purification in hormone-responsive human breast cancer cell nuclei. Proteomics. 2011;11(1):159–65.
Tarallo R, Bamundo A, Nassa G, Nola E, Paris O, Ambrosino C, et al. Identification of proteins associated with ligand-activated estrogen receptor alpha in human breast cancer cell nuclei by tandem affinity purification and nano LC-MS/MS. Proteomics. 2011;11(1):172–9.
Ambrosino C, Tarallo R, Bamundo A, Cuomo D, Franci G, Nassa G, et al. Identification of a hormone-regulated dynamic nuclear actin network associated with estrogen receptor alpha in human breast cancer cell nuclei. Mol Cell Proteomics. 2010;9(6):1352–67.
Nassa G, Tarallo R, Guzzi PH, Ferraro L, Cirillo F, Ravo M, et al. Comparative analysis of nuclear estrogen receptor alpha and beta interactomes in breast cancer cells. Mol Biosyst. 2011;7(3):667–76.
Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336.
Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–6.
Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2014;33(46):5311–8.
Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene. 2015;34(1):1-14.
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science (New York, NY). 2005;309(5740):1559–63.
Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, et al. Cancer-associated regulation of alternative splicing. Nat Struct Mol Biol. 2009;16(6):670–6.
Salama SA, Mohammad MA, Diaz-Arrastia CR, Kamel MW, Kilic GS, Ndofor BT, et al. Estradiol-17beta upregulates pyruvate kinase M2 expression to coactivate estrogen receptor-alpha and to integrate metabolic reprogramming with the mitogenic response in endometrial cells. J Clin Endocrinol Metab. 2014;99(10):3790–9.
Lal S, Allan A, Markovic D, Walker R, Macartney J, Europe-Finner N, et al. Estrogen alters the splicing of type 1 corticotropin-releasing hormone receptor in breast cancer cells. Sci Signal. 2013;6(282):ra53.
Bhat-Nakshatri P, Song EK, Collins NR, Uversky VN, Dunker AK, O’Malley BW, et al. Interplay between estrogen receptor and AKT in estradiol-induced alternative splicing. BMC Med Genomics. 2013;6:21.
Nassa G, Tarallo R, Giurato G, De Filippo MR, Ravo M, Rizzo F, et al. Post-transcriptional regulation of human breast cancer cell proteome by unliganded estrogen receptor beta via microRNAs. Mol Cell Proteomics. 2014;13(4):1076–90.
Shen S, Park JW, Huang J, Dittmar KA, Lu ZX, Zhou Q, et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res. 2012;40(8):e61.
Lang F, Shumilina E. Regulation of ion channels by the serum- and glucocorticoid-inducible kinase SGK1. Faseb J. 2012;27(1):3–12.
Lang F, Stournaras C. Serum and glucocorticoid inducible kinase, metabolic syndrome, inflammation, and tumor growth. Hormones (Athens). 2013;12(2):160–71.
Simon P, Schneck M, Hochstetter T, Koutsouki E, Mittelbronn M, Merseburger A, et al. Differential regulation of serum- and glucocorticoid-inducible kinase 1 (SGK1) splice variants based on alternative initiation of transcription. Cell Physiol Biochem. 2007;20(6):715–28.
Pal S, Gupta R, Kim H, Wickramasinghe P, Baubet V, Showe LC, et al. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 2011;21(8):1260–72.
Shabalina SA, Ogurtsov AY, Spiridonov NA, Koonin EV. Evolution at protein ends: major contribution of alternative transcription initiation and termination to the transcriptome and proteome diversity in mammals. Nucleic Acids Res. 2014;42(11):7132–44.
Schweitzer JK, Sedgwick AE, D’Souza-Schorey C. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin Cell Dev Biol. 2011;22(1):39–47.
Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53.
Cicatiello L, Mutarelli M, Grober OM, Paris O, Ferraro L, Ravo M, et al. Estrogen receptor alpha controls a gene network in luminal-like breast cancer cells comprising multiple transcription factors and microRNAs. Am J Pathol. 2010;176(5):2113–30.
Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39(6):e35.
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45.
Menzl I, Lebeau L, Pandey R, Hassounah NB, Li FW, Nagle R, et al. Loss of primary cilia occurs early in breast cancer development. Cilia. 2014;3:7.
Madak-Erdogan Z, Charn TH, Jiang Y, Liu ET, Katzenellenbogen JA, Katzenellenbogen BS. Integrative genomics of gene and metabolic regulation by estrogen receptors alpha and beta, and their coregulators. Mol Syst Biol. 2013;9:676.
Murray MV, Kobayashi R, Krainer AR. The type 2C Ser/Thr phosphatase PP2Cgamma is a pre-mRNA splicing factor. Genes Dev. 1999;13(1):87–97.
Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4(2):e27.
Han J, Shen Q. Targeting gamma-secretase in breast cancer. Breast cancer (Dove Medical Press). 2012;4:83–90.
Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33.
Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, et al. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12(15):2269–77.
Acar M, Jafar-Nejad H, Takeuchi H, Rajan A, Ibrani D, Rana NA, et al. Rumi is a CAP10 domain glycosyltransferase that modifies Notch and is required for Notch signaling. Cell. 2008;132(2):247–58.
Lu L, Hou X, Shi S, Korner C, Stanley P. Slc35c2 promotes Notch1 fucosylation and is required for optimal Notch signaling in mammalian cells. J Biol Chem. 2010;285(46):36245–54.
Mo JS, Ann EJ, Yoon JH, Jung J, Choi YH, Kim HY, et al. Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by downregulation of protein stability through Fbw7 ubiquitin ligase. J Cell Sci. 2010;124(Pt 1):100–12.
Mizuhara E, Nakatani T, Minaki Y, Sakamoto Y, Ono Y, Takai Y. MAGI1 recruits Dll1 to cadherin-based adherens junctions and stabilizes it on the cell surface. J Biol Chem. 2005;280(28):26499–507.
Reedijk M. Notch signaling and breast cancer. Adv Exp Med Biol. 2012;727:241–57.
Nwabo Kamdje AH, Seke Etet PF, Vecchio L, Muller JM, Krampera M, Lukong KE. Signaling pathways in breast cancer: therapeutic targeting of the microenvironment. Cell Signal. 2014;26(12):2843–56.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36.
Acknowledgements
Work supported by: Italian Association for Cancer Research (Grants IG-13176), Italian Ministries of Education, University and Research (Grants 2010LC747T, RBFR12W5V5_003 and PON03PE_00146_1) and Health (Young Researcher Grants GR-2011-02350476 and GR-2011-02347781), National Research Council (Flagship Project Interomics), the University of Salerno (FARB 2014). G.N. is supported by a ‘Mario e Valeria Rindi’ fellowship of the Italian Foundation for Cancer Research; A.R. is a PhD student of the Research Doctorate in ‘Molecular and Translational Oncology and Innovative Medical-Surgical Technologies’, University of Catanzaro ‘Magna Graecia’; C.S. is supported by the UCLA Scholars in Oncologic Molecular Imaging Fellowship (National Institutes of Health R25 CA098010).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
All authors participated in conception and design of the study and manuscript drafting and revision, GN, MR, FR and RT preformed in vitro experimental work and sequencing, DND, AR, DM and GG performed the statistical and bioinformatic analyses, CS preformed functional investigations on alternatively splices genes, CS and AW coordinated and finalized figure preparation, manuscript drafting and revision. All authors read and approved the final manuscript.
Dougba Noel Dago and Claudio Scafoglio contributed equally to this work.
Additional files
Additional file 1: Figure S1.
Description: KEGG pathway (Kanehisa et al. Nucleic Acids Res 2014, 42:D199) analysis of ERα and ERβ interactors involved in Spliceosome Pathway. Blue and red boxes highlight ERα and ERβ interacting proteins, respectively.
Additional file 2: Tables S1.
A: Ct-ERβ gene list. Genes with a FPKM value ≥0.5, q-value ≤0.05 and |FC| ≥ 1.3 in the Ct-ERβ BC cell line. B: Nt-ERβ gene list. Genes with a FPKM value ≥0.5, q-value ≤0.05 and |FC| ≥1.3 in the Nt-ERβ BC cell line. C: MCF-7 wt gene list. Genes with a FPKM value ≥0.5, q-value ≤0.05 and |FC| ≥1.3 in the wt MCF-7 BC cell line. D: Commonly regulated genes in both ERβ + BC cell lines. Genes consistently regulated in both ERβ + BC cells with a q-value ≤0.05 and |FC| ≥1.3. E: Ct-ERβ isoform list. Isoforms with a FPKM value ≥0.5, q-value ≤0.05 and |FC| ≥1.3 in the Ct-ERβ BC cell line. F: Nt-ERβ isoform list. Isoforms with a FPKM value ≥0.5, q-value ≤0.05 and |FC| ≥1.3 in the Nt-ERβ BC cell line. G: MCF-7 wt isoform list. Isoforms with a FPKM value ≥0.5, FDR ≤0.05 and |FC| ≥1.3 in the wt MCF-7 BC cell line.
Additional file 3: Tables S2.
A: List of exon skipping events in Ct-ERβ, B: List of mutually exclusive exons in Ct-ERβ, C: List of retained introns in Ct-ERβ, D: List of alternative 3’ splice sites in Ct-ERβ, E: List of alternative 5’ splice sites in Ct-ERβ, F: List of exon skipping events in Nt-ERβ, G: List of mutually exclusive exons in Nt-ERβ, H: List of retained introns in Nt-ERβ, I: List of alternative 3’ splice sites in Nt-ERβ, J: List of alternative 5’ splice sites in Nt-ERβ, K: List of exon skipping events in wt MCF-7, L. List of mutually exclusive exons in wt MCF-7, M: List of retained introns in wt MCF-7, N. List of alternative 3’ splice sites in wt MCF-7, O: List of alternative 5’ splice sites in wt MCF-7.
Additional file 4: Table S3.
Discordant Isoforms ERβ+/ERβ-. List of all isoforms with opposite regulation pattern in ERβ+/ERβ-.
Additional file 5: Tables S4.
A: Differential Promoter Usage Ct-ERβ. Gene list with differential promoter usage (DPU) in the Ct-ERβ BC cell line (FDR ≤0.05). B: Differential Promoter Usage Nt-ERβ. Gene list with DPU in the Nt-ERβ BC cell line (FDR ≤0.05). C: Differential Promoter Usage MCF-7 wt. Gene list with DPU in the wt MCF-7 BC cell line (FDR ≤0.05). D: Differential Spliced Gene Ct-ERβ. Gene list differentially spliced (DSGs) in the Ct-ERβ BC cell line (FDR ≤0.05). E: Differential Spliced Gene Nt-ERβ. Gene list differentially spliced (DSGs) in the Nt-ERβ BC cell line (FDR ≤0.05). F: Differential Spliced Gene MCF-7 wt. Gene list differentially spliced (DSGs) in the wt MCF-7 BC cell line (FDR ≤0.05).
Additional file 6: Table S5.
PSD3 TSSs expression values. Transcript starting sites (TSSs) expression value list of the PSD3 gene, expressed in FPKM with relative FC, FDR and p-value in Ct-ERβ, Nt-ERβ and MCF-7 wt BC cell lines.
Additional file 7: Tables S6.
A: ERα binding sites in Ct-ERβ. Gene list of DSGs including ERα binding sites in a 10 kb window around the gene in the Ct-ERβ BC cell line. B: ERβ binding sites in Ct-ERβ. Gene list of DSGs including ERβ binding sites in a 10 kb window around the gene in the Ct-ERβ BC cell line. C: ERα binding sites in Nt-ERβ. Gene list of DSGs including ERα binding sites in a 10 kb window around the gene in the Nt-ERβ BC cell line. D: ERβ binding sites in Nt-ERβ. Gene list of DSGs including ERβ binding sites in a 10 kb window around the gene in the Nt-ERβ BC cell line. E: ERα binding sites in MCF-7 wt. Gene list of DSGs including ERα binding sites in a 10 kb window around the gene in the wt MCF-7 BC cell line.
Additional file 8: Figure S2.
Heat map representing the clustered density matrix of ERα and ERβ binding sites. Chromatin immunoprecipitation of ERα and ERβ in Ct-ERβ and wt MCF-7 cells show different ER binding profiles (highlighted by square brackets). In the clustering, each line represents a genomic location of a binding site with its surrounding ±1.5 kb region. This matrix was subjected to k-means clustering.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver (https://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Dago, D.N., Scafoglio, C., Rinaldi, A. et al. Estrogen receptor beta impacts hormone-induced alternative mRNA splicing in breast cancer cells. BMC Genomics 16, 367 (2015). https://doi.org/10.1186/s12864-015-1541-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-015-1541-1