- Research
- Open access
- Published:
Discovery of novel RNA viruses through analysis of fungi-associated next-generation sequencing data
BMC Genomics volume 25, Article number: 517 (2024)
Abstract
Background
Like all other species, fungi are susceptible to infection by viruses. The diversity of fungal viruses has been rapidly expanding in recent years due to the availability of advanced sequencing technologies. However, compared to other virome studies, the research on fungi-associated viruses remains limited.
Results
In this study, we downloaded and analyzed over 200 public datasets from approximately 40 different Bioprojects to explore potential fungal-associated viral dark matter. A total of 12 novel viral sequences were identified, all of which are RNA viruses, with lengths ranging from 1,769 to 9,516 nucleotides. The amino acid sequence identity of all these viruses with any known virus is below 70%. Through phylogenetic analysis, these RNA viruses were classified into different orders or families, such as Mitoviridae, Benyviridae, Botourmiaviridae, Deltaflexiviridae, Mymonaviridae, Bunyavirales, and Partitiviridae. It is possible that these sequences represent new taxa at the level of family, genus, or species. Furthermore, a co-evolution analysis indicated that the evolutionary history of these viruses within their groups is largely driven by cross-species transmission events.
Conclusions
These findings are of significant importance for understanding the diversity, evolution, and relationships between genome structure and function of fungal viruses. However, further investigation is needed to study their interactions.
Introduction
Viruses are among the most abundant and diverse biological entities on Earth; they are ubiquitous in the natural environment but difficult to culture and detect [1,2,3]. In recent decades, the significant advancements in omics have transformed the field of virology and enabled researchers to detect potential viruses in a variety of environmental samples, helping us to expand the known diversity of viruses and explore the “dark matter” of viruses that may exist in vast quantities [4]. In most cases, the hosts of these newly discovered viruses exhibit only asymptomatic infections [5, 6], and they even play an important role in maintaining the balance, stability, and sustainable development of the biosphere [7]. But some viruses may be involved in the emergence and development of animal or plant diseases. For example, the tobacco mosaic virus (TMV) causes poor growth in tobacco plants, while norovirus is known to cause diarrhea in mammals [8, 9]. In the field of fungal research, viral infections have significantly reduced the yield of edible fungi, thereby attracting increasing attention to fungal diseases caused by viruses [10]. However, due to their apparent relevance to health [11], fungal-associated viruses have been understudied compared to viruses affecting humans, animals, or plants.
Mycoviruses (also known as fungal viruses) are widely distributed in various fungi and fungal-like organisms [12]. The first mycoviruses were discovered in the 1960s by Hollings M in the basidiomycete Agaricus bisporus, an edible cultivated mushroom [13]. Shortly thereafter, Ellis LF et al. reported mycoviruses in the ascomycete Penicillium stoloniferum, confirming that viral dsRNA is responsible for interferon stimulation in mammals [13,14,15]. In recent years, the diversity of known mycoviruses has rapidly increased with the development and widespread application of sequencing technologies [16,17,18,19,20]. According to the classification principles of the International Committee for the Taxonomy of Viruses (ICTV), mycoviruses are currently classified into 24 taxa, consisting of 23 families and 1 genus (Botybirnavirus) [21]. Most mycoviruses belong to double-stranded (ds) RNA viruses, such as families Totiviridae, Partitiviridae, Reoviridae, Chrysoviridae, Megabirnaviridae, Quadriviridae, and genus Botybirnavirus, or positive-sense single-stranded (+ ss) RNA viruses, such as families Alphaflexiviridae, Gammaflexiviridae, Barnaviridae, Hypoviridae, Endornaviridae, Metaviridae and Pseudoviridae. However, negative-sense single-stranded (-ss) RNA viruses (family Mymonaviridae) and single-stranded (ss) DNA viruses (family Genomoviridae) have also been described [22]. The taxonomy of mycoviruses is continually refined as novel mycoviruses that cannot be classified into any established taxon are identified. While the vast majority of fungi-infecting viruses do not show infection characteristics and have no significant impact on their hosts, some mycoviruses have inhibitory effects on the phenotype of the host, leading to hypovirulence in phytopathogenic fungi [23]. The use of environmentally friendly, low-virulence-related mycoviruses such as Chryphonectria hypovirus 1 (CHV-1) for biological control has been considered a viable alternative to chemical fungicides [24]. With the deepening of research, an increasing number of mycoviruses that can cause fungal phenotypic changes have been identified [3, 23, 25]. Therefore, understanding the distribution of these viruses and their effects on hosts will allow us to determine whether their infections can be prevented and treated.
To explore the viral dark matter hidden within fungi, this study collected over 200 available fungal-associated libraries from approximately 40 Bioprojects in the Sequence Read Archive (SRA) database, uncovering novel RNA viruses within them. We further elucidated the genetic relationships between known viruses and these newfound ones, thereby expanding our understanding of fungal-associated viruses and providing assistance to viral taxonomy.
Materials and methods
Genome assembly
To discover novel fungal-associated viruses, we downloaded 236 available libraries from the SRA database, corresponding to 32 fungal species (Supplementary Table 1). Pfastq-dump v0.1.6 (https://github.com/inutano/pfastq-dump) was used to convert SRA format files to fastq format files. Subsequently, Bowtie2 v2.4.5 [26] was employed to remove host sequences. Primer sequences of raw reads underwent trimming using Trim Galore v0.6.5 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore), and the resulting files underwent quality control with the options ‘–phred33 –length 20 –stringency 3 –fastqc’. Duplicated reads were marked using PRINSEQ-lite v0.20.4 (-derep 1). All SRA datasets were then assembled in-house pipeline. Paired-end reads were assembled using SPAdes v3.15.5 [27] with the option ‘-meta’, while single-end reads were assembled with MEGAHIT v1.2.9 [28], both using default parameters. The results were then imported into Geneious Prime v2022.0.1 (https://www.geneious.com) for sorting and manual confirmation. To reduce false negatives during sequence assembly, further semi-automatic assembly of unmapped contigs and singlets with a sequence length < 500 nt was performed. Contigs with a sequence length > 1,500 nt after reassembly were retained. Individual contigs were then used as references for mapping to the raw data using the Low Sensitivity/Fastest parameter in Geneious Prime. In addition, mixed assembly was performed using MEGAHIT in combination with BWA v0.7.17 [29] to search for unused reads that might correspond to low-abundance contigs.
Searching for novel viruses in fungal libraries
We identified novel viral sequences present in fungal libraries through a series of steps. To start, we established a local viral database, consisting of the non-redundant protein (nr) database downloaded in August 2023, along with IMG/VR v3 [30], for screening assembled contigs. The contigs labeled as “viruses” and exhibiting less than 70% amino acid (aa) sequence identity with the best match in the database were imported into Geneious Prime for manual mapping. Putative open reading frames (ORFs) were predicted by Geneious Prime using built-in parameters (Minimum size: 100) and were subsequently verified by comparison to related viruses. The annotations of these ORFs were based on comparisons to the Conserved Domain Database (CDD). The sequences after manual examination were subjected to genome clustering using MMseqs2 (-k 0 -e 0.001 –min-seq-id 0.95 -c 0.9 –cluster-mode 0) [31]. After excluding viruses with high aa sequence identity (> 70%) to known viruses, a dataset containing a total of 12 RNA viral sequences was obtained. The non-redundant fungal virus dataset was compared against the local database using the BLASTx program built in DIAMOND v2.0.15 [32], and significant sequences with a cut-off E-value of < 10–5 were selected. The coverage of each sequence in all libraries was calculated using the pileup tool in BBMap. Taxonomic identification was conducted using TaxonKit [33] software, along with the rma2info program integrated into MEGAN6 [34]. The RNA secondary structure prediction of the novel viruses was conducted using RNA Folding Form V2.3 (http://www.unafold.org/mfold/applications/rna-folding-form-v2.php).
Phylogenetic analysis
To infer phylogenetic relationships, nucleotide and their encoded protein sequences of reference strains belonging to different groups of corresponding viruses were downloaded from the NCBI GenBank database, along with sequences of proposed species pending ratification. Related sequences were aligned using the alignment program within the CLC Genomics Workbench 10.0, and the resulting alignment was further optimized using MUSCLE in MEGA-X [35]. Sites containing more than 50% gaps were temporarily removed from the alignments. Maximum-likelihood (ML) trees were then constructed using IQ-TREE v1.6.12 [36]. All phylogenetic trees were created using IQ-TREE with 1,000 bootstrap replicates (-bb 1000) and the ModelFinder function (-m MFP). Interactive Tree Of Life (iTOL) was used for visualizing and editing phylogenetic trees [37]. Colorcoded distance matrix analysis between novel viruses and other known viruses were performed with Sequence Demarcation Tool v1.2 [38].
To illustrate cross-species transmission and co-divergence between viruses and their hosts across different virus groups, we reconciled the co-phylogenetic relationships between these viruses and their hosts. The evolutionary tree and topologies of the hosts involved in this study were obtained from the TimeTree [39] website by inputting their Latin names. The viruses in the phylogenetic tree for which the host cannot be recognized through published literature or information provided by the authors are disregarded. The co-phylogenetic plots (or ‘tanglegram’) generated using the R package phytools [40] visually represent the correspondence between host and virus trees, with lines connecting hosts and their respective viruses. The event-based program eMPRess [41] was employed to determine whether the pairs of virus groups and their hosts undergo coevolution. This tool reconciles pairs of phylogenetic trees according to the Duplication-Transfer-Loss (DTL) model [42], employing a maximum parsimony formulation to calculate the cost of each coevolution event. The cost of duplication, host-jumping (transfer), and extinction (loss) event types were set to 1.0, while host-virus co-divergence was set to zero, as it was considered the null event.
Data availability
The data reported in this paper have been deposited in the GenBase in National Genomics Data Center [43], Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession numbers C_AA066339.1-C_AA066350.1 that are publicly accessible at https://ngdc.cncb.ac.cn/genbase. Please refer to Table 1 for details.
Results
Twelve novel RNA viruses associated with fungi
We investigated fungi-associated novel viruses by mining publicly available metagenomic and transcriptomic fungal datasets. In total, we collected 236 datasets, which were categorized into four fungal phyla: Ascomycota (159), Basidiomycota (47), Chytridiomycota (15), and Zoopagomycota (15). These phyla corresponded to 20, 8, 2, and 2 different fungal genera, respectively (Supplementary Table 1). A total of 12 sequences containing complete coding DNA sequences (CDS) for RNA-dependent RNA polymerase (RdRp) have been identified, ranging in length from 1,769 nt to 9,516 nt. All of these sequences have less than 70% aa identity with RdRp sequences from any currently known virus (ranging from 32.97% to 60.43%), potentially representing novel families, genera, or species (Table 1). Some of the identified sequences were shorter than the reference genomes of RNA viruses, suggesting that these viral sequences represented partial sequences of viral genomes. To exclude the possibility of transient viral infections in hosts or de novo assembly artefacts in co-infection detection, we extracted the nucleotide sequences of the coding regions of these 12 sequences and mapped them to all collected libraries to compute coverage (Supplementary Table 2). The results revealed varying degrees of read matches for these viral genomes across different libraries, spanning different fungal species. Although we only analyzed sequences longer than 1,500 nt, it is worth noting that we also discovered other viral reads in many libraries. However, we were unable to assemble them into sufficiently long contigs, possibly due to library construction strategies or sequencing depth. In any case, this preliminary finding reveals a greater diversity of fungal-associated viruses than previously considered.
Positive-sense single-stranded RNA viruses
(i) Mitoviridae
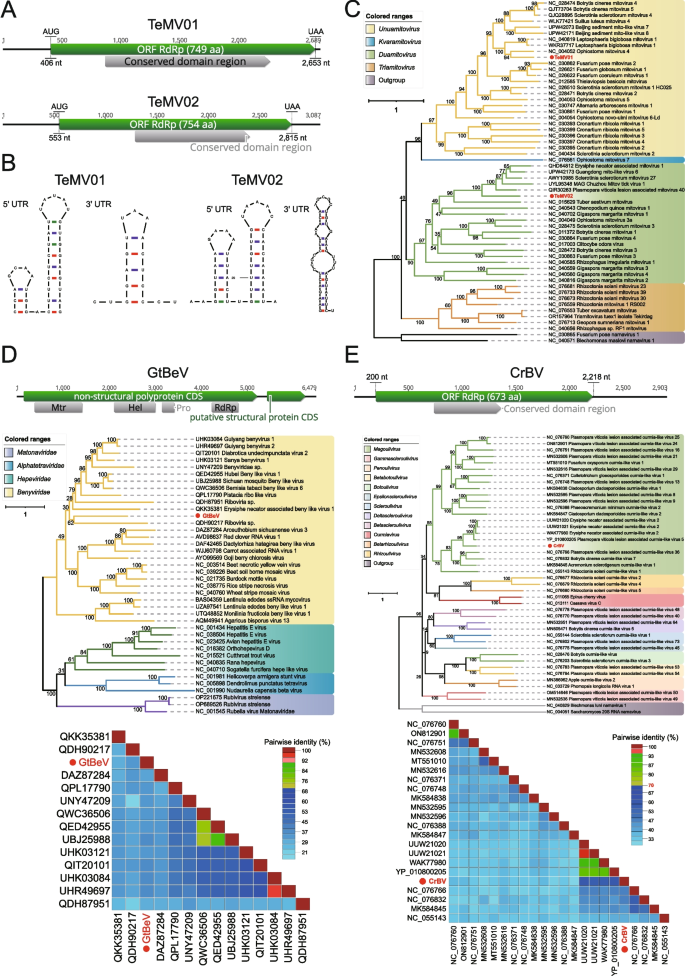
Members of the family Mitoviridae (order Cryppavirales) are monopartite, linear, positive-sense ( +) single-stranded (ss) RNA viruses with genome size of approximately 2.5–2.9 kb [44], carrying a single long open reading frame (ORF) which encodes a putative RdRp. Mitoviruses have no true virions and no structural proteins, virus genome is transmitted horizontally through mating or vertically from mother to daughter cells [45]. They use mitochondria as their sites of replication and have typical 5' and 3' untranslated regions (UTRs) of varying sizes, which are responsible for viral translation and replicase recognition [46]. According to the taxonomic principles of ICTV, the viruses belonging to the family Mitoviridae are divided into four genera, namely Duamitovirus, Kvaramitovirus, Triamitovirus and Unuamitovirus. In this study, two novel viruses belonging to the family Mitoviridae were identified in the same library (SRR12744489; Species: Thielaviopsis ethacetica), named Thielaviopsis ethacetica mitovirus 1 (TeMV01) and Thielaviopsis ethacetica mitovirus 2 (TeMV02), respectively (Fig. 1A). The genome sequence of TeMV01 spans 2,689 nucleotides in length with a GC content of 32.2%. Its 5' and 3' UTRs comprise 406 nt and 36 nt, respectively. Similarly, the genome sequence of TeMV02 extends 3,087 nucleotides in length with a GC content of 32.6%. Its 5' and 3' UTRs consist of 553 and 272 nt, respectively. The 5' and 3' ends of both genomes are predicted to have typical stem-loop structures (Fig. 1B). In order to determine the evolutionary relationship between these two mitoviruses and other known mitoviruses, phylogenetic analysis based on RdRp showed that viral strains were divided into 2 genetic lineages in the genera Duamitovirus and Unuamitovirus (Fig. 1C). In the genus Unuamitovirus, TeMV01 was clustered with Ophiostoma mitovirus 4, exhibiting the highest aa identity of 51.47%, while in the genus Duamitovirus, TeMV02 was clustered with a strain isolated from Plasmopara viticola, showing the highest aa identity of 42.82%. According to the guidelines from the ICTV regarding the taxonomy of the family Mitoviridae, a species demarcation cutoff of < 70% aa sequence identity is established [47]. Drawing on this recommendation and phylogenetic inferences, these two viral strains could be presumed to be novel viral species [48].
Identification of novel positive-sense single-stranded RNA viruses in fungal sequencing libraries. A Genome organization of two novel mitoviruses; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. B Predicted RNA secondary structures of the 5'- and 3'-terminal regions. C ML phylogenetic tree of members of the family Mitoviridae. The best-fit model (LG + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font. D The genome organization of GtBeV is depicted at the top; in the middle is the ML phylogenetic tree of members of the family Benyviridae. The best-fit model (VT + F + R5) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. At the bottom is the distance matrix analysis of GeBeV identified in Gaeumannomyces tritici. Pairwise sequence comparison produced with the RdRp amino acid sequences within the ML tree. E The genome organization of CrBV is depicted at the top; in the middle is the ML phylogenetic tree of members of the family Botourmiaviridae. The best-fit model (VT + F + R5) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. At the bottom is the distance matrix analysis of CrBV identified in Clonostachys rosea. Pairwise sequence comparison produced with the RdRp amino acid sequences within the ML tree
(ii) Benyviridae
The family Benyviridae is comprised of multipartite plant viruses that are rod-shaped, approximately 85–390 nm in length and 20 nm in diameter. Within this family, there is a single genus, Benyvirus [49]. It is reported that one species within this genus,Beet necrotic yellow vein virus, can cause widespread and highly destructive soil-borne ‘rhizomania’ disease of sugar beet [50]. A full-length RNA1 sequence related to Benyviridae has been detected from Gaeumannomyces tritici (ERR3486062), with a length of 6,479 nt. It possesses a poly(A) tail at the 3' end and is temporarily designated as Gaeumannomyces tritici benyvirus (GtBeV). BLASTx results indicate a 34.68% aa sequence identity with the best match found (Fig. 1D). The non-structural polyprotein CDS of RNA1 encodes a large replication-associated protein of 1,688 amino acids with a molecular mass of 190 kDa. Four domains were predicted in this polyprotein corresponding to representative species within the family Benyviridae. The viral methyltransferase (Mtr) domain spans from nucleotide position 386 to 1411, while the RNA helicase (Hel) domain occupies positions 2113 to 2995 nt. Additionally, the protease (Pro) domain is located between positions 3142 and 3410 nt, and the RdRp domain is located at 4227 to 4796 nt. A phylogenetic analysis was conducted by integrating RdRp sequences of viruses closely related to GtBeV. The result revealed that GtBeV clustered within the family Benyviridae, exhibiting substantial evolutionary divergence from any other sequences. Consequently, this virus likely represents a novel species in the family Benyviridae.
(iii) Botourmiaviridae
The family Botourmiaviridae comprises viruses infecting plants and filamentous fungi, which may possess mono- or multi-segmented genomes [51]. Recent research has led to a rapid expansion in the number of viruses within the family Botourmiaviridae, increasing from the confirmed 4 genera in 2020 to a total of 12 genera. A contig identified from Clonostachys rosea (ERR5928658) using the BLASTx method exhibited similarity to viruses in the family Botourmiaviridae. After manual mapping, a 2,903 nt-long genome was obtained, tentatively named Clonostachys rosea botourmiavirus (CrBV), which includes a complete RdRP region (Fig. 1E). Based on phylogenetic analysis using RdRp, CrBV clustered with members of the genus Magoulivirus, sharing 56.58% aa identity with a strain identified from Eclipta prostrata. However, puzzlingly, according to the ICTV's Genus/Species demarcation criteria, members of different genera/species within the family Botourmiaviridae share less than 70%/90% identity in their complete RdRP amino acid sequences. Furthermore, the RdRp sequences with accession numbers NC_055143 and NC_076766, both considered to be members of the genus Magoulivirus, exhibited only 39.05% aa identity to each other. Therefore, CrBV should at least be considered as a new species within the family Botourmiaviridae.
(iv) Deltaflexiviridae
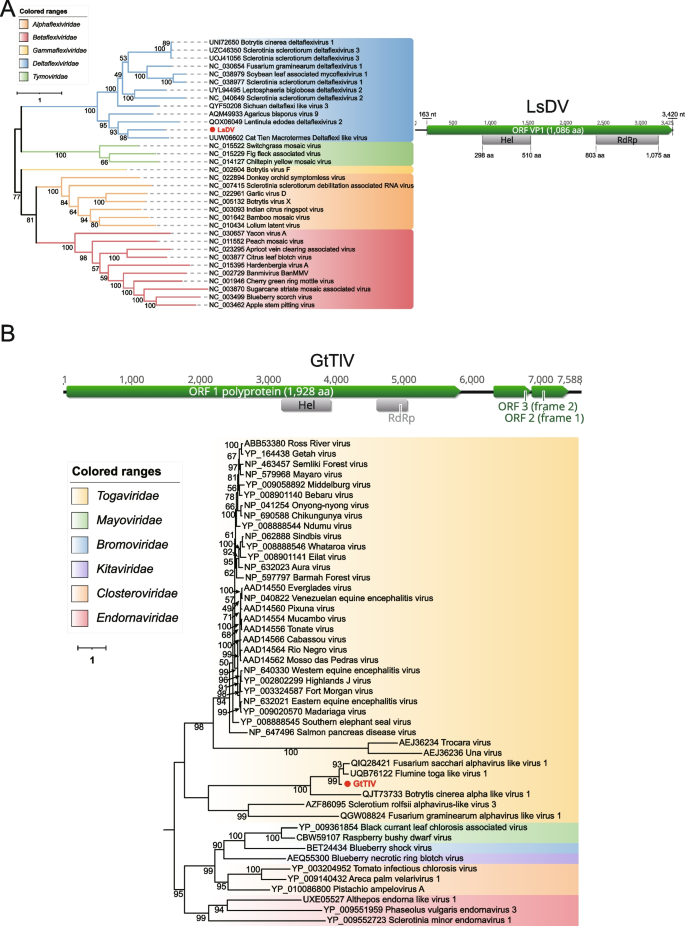
An assembled sequence of 3,425 nucleotides in length Lepista sordida deltaflexivirus (LsDV), derived from Lepista sordida (DRR252167) and showing homology to Deltaflexiviridae within the order Tymovirales, was obtained. The Tymovirales comprises five recognized families: Alphaflexiviridae, Betaflexiviridae, Deltaflexiviridae, Gammaflexiviridae, and Tymoviridae [52]. The Deltaflexiviridae currently only includes one genus, the fungal-associated deltaflexivirus; they are mostly identified in fungi or plants pathogens [53]. LsDV was predicted to have a single large ORF, VP1, which starts with an AUG codon at nt 163–165 and ends with a UAG codon at nt 3,418–3,420. This ORF encodes a putative polyprotein of 1,086 aa with a calculated molecular mass of 119 kDa. Two conserved domains within the VP1 protein were identified: Hel and RdRp (Fig. 2A). However, the Mtr was missing, indicating that the 5' end of this polyprotein is incomplete. According to the phylogenetic analysis of RdRp, LsDV was closely related to viruses of the family Deltaflexiviridae and shared 46.61% aa identity with a strain (UUW06602) isolated from Macrotermes carbonarius. Despite this, according to the species demarcation criteria proposed by ICTV, because we couldn't recover the entire replication-associated polyprotein, LsDV cannot be regarded as a novel species at present.
Identification of novel members of family Deltaflexiviridae and Toga-like virus in fungal sequencing libraries. A On the right side of the image is the genome organization of LsDV; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. ML phylogenetic tree of members of the family Deltaflexiviridae. The best-fit model (VT + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font. B The genome organization of GtTlV is depicted at the top; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. ML phylogenetic tree of members of the order Martellivirales. The best-fit model (LG + R7) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font
(v) Toga-like virus
Members of the family Togaviridae are primarily transmitted by arthropods and can infect a wide range of vertebrates, including mammals, birds, reptiles, amphibians, and fish [54]. Currently, this family only contains a single confirmed genus, Alphavirus. A contig was discovered in Gaeumannomyces tritici (ERR3486058), it is 7,588 nt in length with a complete ORF encoding a putative protein of 1,928 aa, which had 60.43% identity to Fusarium sacchari alphavirus-like virus 1 (QIQ28421) with 97% coverage. Phylogenetic analysis showed that it did not cluster with classical alphavirus members such as VEE, WEE, EEE, SF complex [54], but rather with several sequences annotated as Toga-like that were available (Fig. 2B). It was provisionally named Gaeumannomyces tritici toga-like virus (GtTIV). However, we remain cautious about the accuracy of these so-called Toga-like sequences, as they show little significant correlation with members of the order Martellivirales.
Negative-sense single-stranded RNA viruses
(i) Mymonaviridae
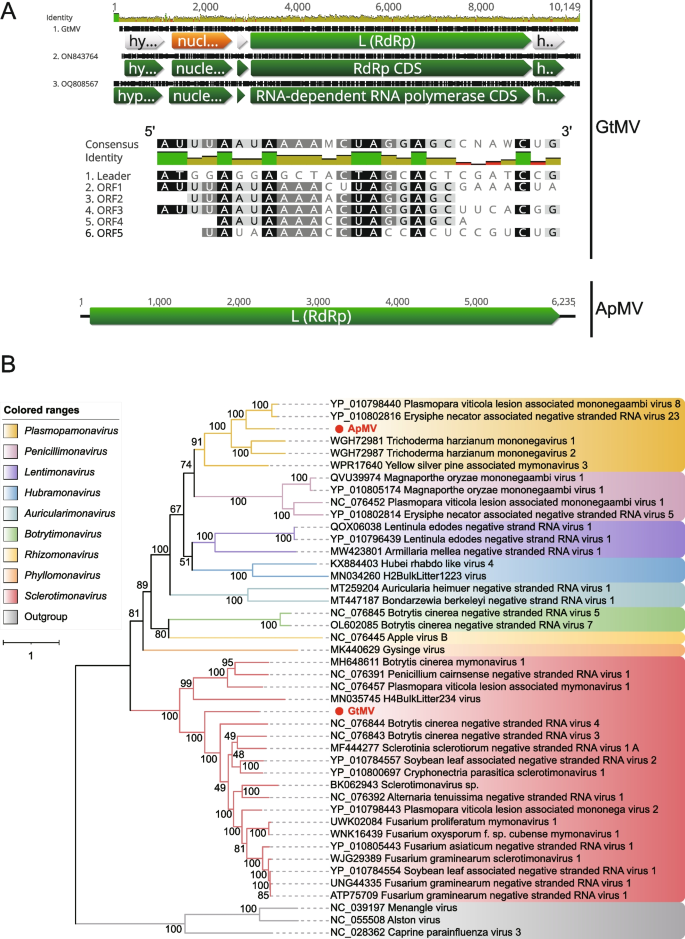
Mymonaviridae is a family of linear, enveloped, negative-stranded RNA genomes in the order Mononegavirales, which infect fungi. They are approximately 10 kb in size and encode six proteins [55]. The famliy Mymonaviridae was established to accommodate Sclerotinia sclerotiorum negative-stranded RNA virus 1 (SsNSRV-1), a novel virus discovered in a hypovirulent strain of Sclerotinia sclerotiorum [56]. According to the ICTV, the family Mymonaviridae currently includes 9 genera, namely Auricularimonavirus, Botrytimonavirus, Hubramonavirus, Lentimonavirus, Penicillimonavirus, Phyllomonavirus, Plasmopamonavirus, Rhizomonavirus and Sclerotimonavirus. Two sequences originating from Gaeumannomyces tritici (ERR3486068) and Aspergillus puulaauensis (DRR266546), respectively, and associated with the family Mymonaviridae, have been identified and provisionally named Gaeumannomyces tritici mymonavirus (GtMV) and Aspergillus puulaauensis mymonavirus (ApMV). GtMV is 9,339 nt long with a GC content of 52.8%. It was predicted to contain 5 discontinuous ORFs, with the largest one encoding RdRp. Additionally, a nucleoprotein and three hypothetical proteins with unknown function were also predicted. A multiple alignment of nucleotide sequences among these ORFs identified a semi-conserved sequence, 5'-UAAAA-CUAGGAGC-3', located downstream of each ORF (Fig. 3A). These regions are likely gene-junction regions in the GtMV genome, a characteristic feature shared by mononegaviruses [57, 58]. For ApMV, a complete RdRp CDS with a length of 1,978 aa was predicted. The BLASTx searches showed that GtMV shared 45.22% identity with the RdRp of Soybean leaf-associated negative-stranded RNA virus 2 (YP_010784557), while ApMV shared 55.90% identity with the RdRp of Erysiphe necator associated negative-stranded RNA virus 23 (YP_010802816). The representative members of the family Mymonaviridae were included in the phylogenetic analysis. The results showed that GtMV and ApMV clustered closely with members of the genera Sclerotimonavirus and Plasmopamonavirus, respectively (Fig. 3B). Members of the genus Plasmopamonavirus are about 6 kb in size and encode for a single protein. Therefore, GtMV and ApMV should be considered as representing new species within their respective genera.
Identification of two new members in the family Mymonaviridae. A At the top is the nucleotide multiple sequence alignment result of GtMV with the reference genomes. the putative ORF for the viral RdRp is depicted by a green box, the predicted nucleoprotein is displayed in a yellow box, and three hypothetical proteins are displayed in gray boxes. The comparison of putative semi-conserved regions between ORFs in GtMV is displayed in the 5' to 3' orientation, with conserved sequences are highlighted. At the bottom is the genome organization of AmPV; the putative ORF for the viral RdRp is depicted by a green box. B ML phylogenetic tree of members of the family Mymonaviridae. The best-fit model (LG + F + R6) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font
(ii) Bunyavirales
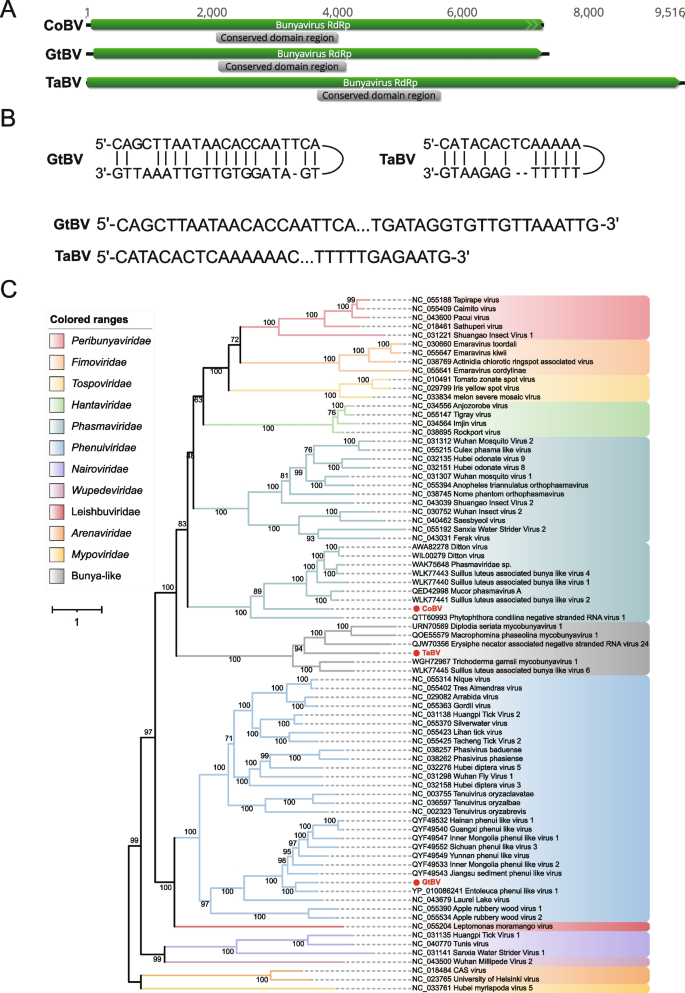
The Bunyavirales (the only order in the class Ellioviricetes) is one of the largest groups of segmented negative-sense single-stranded RNA viruses with mainly tripartite genomes [59], which includes many pathogenic strains that infect arthropods(such as mosquitoes, ticks, sand flies), plants, protozoans, and vertebrates, and even cause severe human diseases. Order Bunyavirales consists of 14 viral families, including Arenaviridae, Cruliviridae, Discoviridae, Fimoviridae, Hantaviridae, Leishbuviridae, Mypoviridae, Nairoviridae, Peribunyaviridae, Phasmaviridae, Phenuiviridae, Tospoviridae, Tulasviridae and Wupedeviridae. In this study, three complete or near complete RNA1 sequences related to bunyaviruses were identified and named according to their respective hosts: CoBV (Conidiobolus obscurus bunyavirus; SRR6181013; 7,277 nt), GtBV (Gaeumannomyces tritici bunyavirus; ERR3486069; 7,364 nt), and TaBV (Thielaviopsis aethacetica bunyavirus; SRR12744489; 9,516 nt) (Fig. 4A). The 5' and 3' terminal RNA segments of GtBV and TaBV complement each other, allowing the formation of a panhandle structure [60], which plays an essential role as promoters of genome transcription and replication [61], except for CoBV, as the 3' terminal of CoBV has not been fully obtained (Fig. 4B). BLASTx results indicated that these three viruses had identities ranging from 32.97% to 54.20% to the best matches in the GenBank database. Phylogenetic analysis indicated that CoBV was classified into the family Phasmaviridae, with distant relationships to any of its genera; GtBV clustered well with members of the genus Entovirus of family Phenuiviridae; while TaBV did not cluster with any known members of families within Bunyavirales, hence provisionally placed within the Bunya-like group (Fig. 4C). Therefore, these three sequences should be considered as potential new family, genus, or species within the order Bunyavirales.
Identification of three new members in the order Bunyavirales. A The genome organization of CoBV, GtBV, and TaBV; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. B The complementary structures formed at the 5' and 3' ends of GtBV and TaBV. C ML phylogenetic tree of members of the order Bunyavirales. The best-fit model (VT + F + R8) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified viruses represented in red font
Double-stranded RNA viruses
Partitiviridae
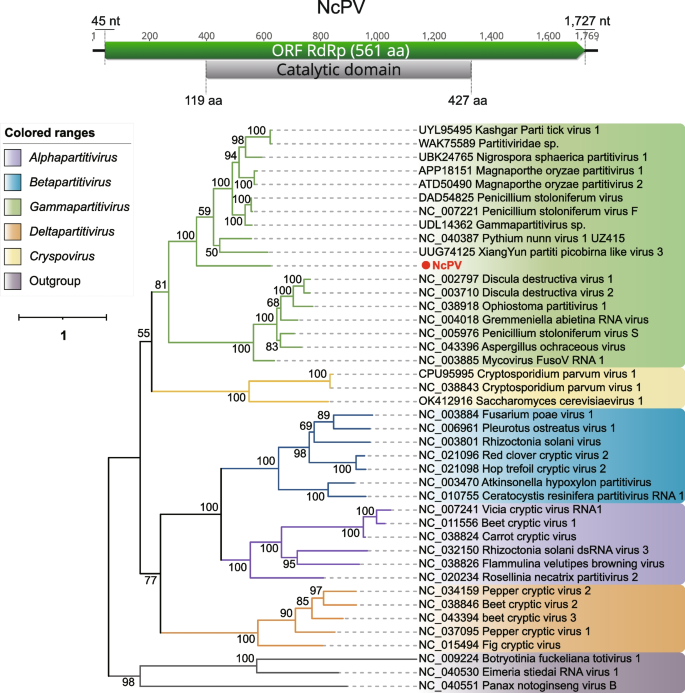
The Partitiviridae is a family of small, non-enveloped viruses, approximately 35–40 nm in diameter, with bisegmented double-stranded (ds) RNA genomes. Each segment is about 1.4–3.0 kb in size, resulting in a total size about 4 kb [62]. The family Partitiviridae is now divided into five genera: Alphapartitivirus, Betapartiivirus, Cryspovirus, Deltapartitivirus and Gammapartitivirus. Each genus has characteristic hosts: plants or fungi for Alphapartitivirus and Betapartitivirus, fungi for Gammapartitivirus, plants for Deltapartitivirus, and protozoa for Cryspovirus [62]. A complete dsRNA1 sequence Neocallimastix californiae partitivirus (NcPV) retrieved from Neocallimastix californiae (SRR15362281) has been identified as being associated with the family Partitiviridae. The BLASTp result indicated that it shared the highest aa identity of 41.5% with members of the genus Gammapartitivirus. According to the phylogenetic tree constructed based on RdRp, NcPV was confirmed to fall within the genus Gammapartitivirus (Fig. 5). Typical members of the genus Gammapartitivirus have two segments in their complete genome, namely dsRNA1 and dsRNA2, encoding RdRp and coat protein, respectively [62]. The larger dsRNA1 segment of NcPV measures 1,769 nt in length, with a GC content of 35.8%. It contains a single ORF encoding a 561 aa RdRp. A CDD search revealed that the RdRp of NcPV harbors a catalytic region spanning from 119 to 427aa. Regrettably, only the complete dsRNA1 segment was obtained. According to the classification principles of ICTV, due to the lack of information regarding dsRNA2, we are unable to propose it as a new species. It is worth noting that according to the Genus demarcation criteria (https://ictv.global/report/chapter/partitiviridae/partitiviridae), members of the genus Gammapartitivirus should have a dsRNA1 length ranging from 1645 to 1787 nt, and the RdRp length should fall between 519 and 539 aa. However, the length of dsRNA1 in NcPV is 1,769 nt, with RdRp being 561 aa, challenging this classification criterion. In fact, multiple strains have already exceeded this criterion, such as GenBank accession numbers: WBW48344, UDL14336, QKK35392, among others.
Identification of a new member in the family Partitiviridae. The genome organization of NcPV is depicted at the top; the putative ORF for the viral RdRp is depicted by a green box, and the predicted conserved domain region is displayed in a gray box. At the bottom is the ML phylogenetic tree of members of the family Partitiviridae. The best-fit model (VT + F + R4) was estimated using IQ-Tree model selection. The bootstrap value is shown at each branch, with the newly identified virus represented in red font
Long-term evolutionary relationships between fungal-associated viruses and hosts
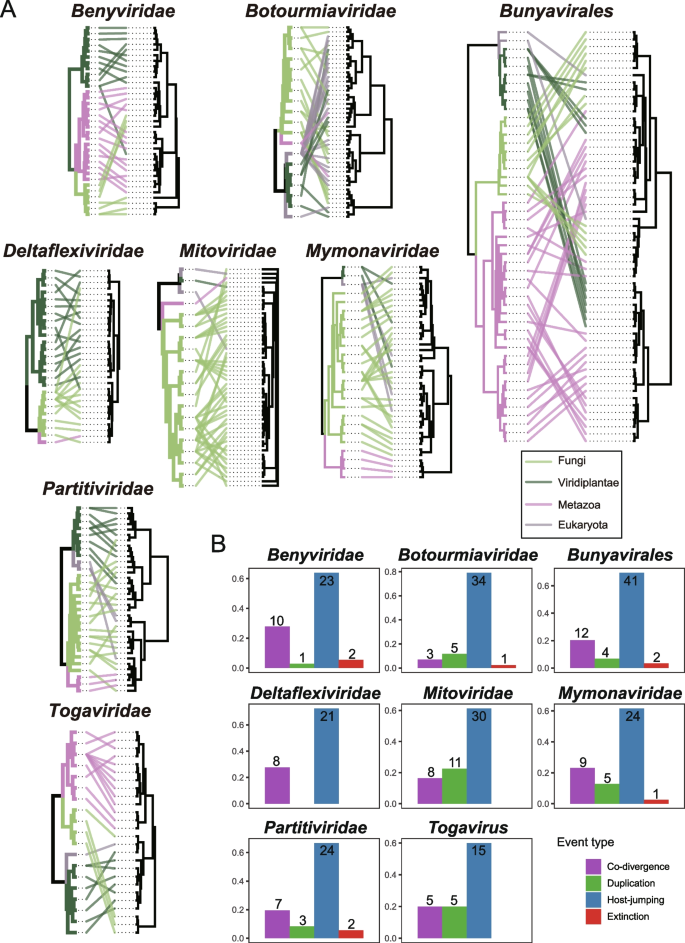
Understanding the co-divergence history between viruses and hosts helps reveal patterns of virus transmission and infection and influences the biodiversity and stability of ecosystems. To explore the frequency of cross-species transmission and co-divergence among fungi-associated viruses, we constructed tanglegrams illustrating the interconnected evolutionary histories of viral families and their respective hosts through phylogenetic trees (Fig. 6A). The results indicated that cross-species transmission (Host-jumping) consistently emerged as the most frequent evolutionary event among all groups of RNA viruses examined in this study (median, 66.79%; range, 60.00% to 79.07%) (Fig. 6B). This finding is highly consistent with the evolutionary patterns of RNA viruses recently identified by Mifsud et al. in their extensive transcriptome survey of plants [63]. Members of the families Botourmiaviridae (79.07%) and Deltaflexiviridae (72.41%) were most frequently involved in cross-species transmission. The frequencies of co-divergence (median, 20.19%; range, 6.98% to 27.78%), duplication (median, 10.60%; range, 0% to 22.45%), and extinction (median, 2.42%; range, 0% to 5.56%) events involved in the evolution of fungi-associated viruses gradually decrease. Specifically, members of the family Benyviridae exhibited the highest frequency of co-divergence events, which also supports the findings reported by Mifsud et al.; certain studies propose that members of Benyviridae are transmitted via zoospores of plasmodiophorid protist [64]. It's speculated that the ancestor of these viruses underwent interkingdom horizontal transfer between plants and protists over evolutionary timelines [65]. Members of the family Mitoviridae showed the highest frequency of duplication events; and members of the families Benyviridae and Partitiviridae demonstrated the highest frequency of extinction events. Not surprisingly, this result is influenced by the current limited understanding of virus-host relationships. On one hand, viruses whose hosts cannot be recognized through published literature or information provided by authors have been overlooked. On the other hand, the number of viruses recorded in reference databases represents just the tip of the iceberg within the entire virosphere. The involvement of a more extensive sample size in the future should change this evolutionary landscape.
Co-evolutionary analysis of virus and host. A Tanglegram of phylogenetic trees for virus orders/families and their hosts. Lines and branches are color-coded to indicate host clades. The cophylo function in phytools was employed to enhance congruence between the host (left) and virus (right) phylogenies. B Reconciliation analysis of virus groups. The bar chart illustrates the proportional range of possible evolutionary events, with the frequency of each event displayed at the top of its respective column
Discussion
Our understanding of the interactions between fungi and their associated viruses has long been constrained by insufficient sampling of fungal species. Advances in metagenomics in recent decades have led to a rapid expansion of the known viral sequence space, but it is far from saturated. The diversity of hosts, the instability of the viral structures (especially RNA viruses), and the propensity to exchange genetic material with other host viruses all contribute to the unparalleled diversity of viral genomes [66]. Fungi are diverse and widely distributed in nature and are closely related to humans. A few fungi can parasitize immunocompromised humans, but their adverse effects are limited. As decomposers in the biological chain, fungi can decompose the remains of plants and animals and maintain the material cycle in the biological world [67]. In agricultural production, many fungi are plant pathogens, and about 80% of plant diseases are caused by fungi. However, little is currently known about the diversity of mycoviruses and how these viruses affect fungal phenotypes, fungal-host interactions, and virus evolution, and the sequencing depth of fungal libraries in most public databases only meets the needs of studying bacterial genomes. Sampling viruses from a larger diversity of fungal hosts should lead to new and improved evolutionary scenarios.
RNA viruses are widespread in deep-sea sediments [68], freshwater [69], sewage [70], and rhizosphere soils [71]. Compared to DNA viruses, RNA viruses are less conserved, prone to mutation, and can transfer between different hosts, potentially forming highly differentiated and unrecognized novel viruses. This characteristic increases the difficulty of monitoring these viruses. Previously, all discovered mycoviruses were RNA viruses. Until 2010, Yu et al. reported the discovery of a DNA virus, namely SsHADV-1, in fungi for the first time [72]. Subsequently, new fungal-related DNA viruses are continually being identified [73,74,75]. Currently, viruses have been found in all major groups of fungi, and approximately 100 types of fungi can be infected by viruses, instances exist where one virus can infect multiple fungi, or one fungus can be infected by several viruses simultaneously. The transmission of mycoviruses differs from that of animal and plant viruses and is mainly categorized into vertical and horizontal transmission [76]. Vertical transmission refers to the spread of the mycovirus to the next generation through the sexual or asexual spores of the fungus, while horizontal transmission refers to the spread of the mycovirus from one strain to another through fusion between hyphae. In the phylum Ascomycota, mycoviruses generally exhibit a low ability to transmit vertically through ascospores, but they are commonly transmitted vertically to progeny strains through asexual spores [77].
In this study, we identified two novel species belonging to different genera within the family Mitoviridae. Interestingly, they both simultaneously infect the same fungus—Thielaviopsis ethacetica, the causal agent of pineapple sett rot disease in sugarcane [78]. Previously, a report identified three different mitoviruses in Fusarium circinatum [79]. These findings suggest that there may be a certain level of adaptability or symbiotic relationship among members of the family Mitoviridae. Benyviruses are typically considered to infect plants, but recent evidence suggests that they can also infect fungi, such as Agaricus bisporus [80], further reinforced by the virus we discovered in Gaeumannomyces tritici. Moreover, members of the family Botourmiaviridae commonly exhibit a broad host range, with viruses closely related to CrBV capable of infecting members of Eukaryota, Viridiplantae, and Metazoa, in addition to fungi (Supplementary Fig. 1). The LsDV identified in this study shared the closest phylogenetic relationship with a virus identified from Macrotermes carbonarius in southern Vietnam (17_N1 + N237) [81]. M. carbonarius is an open-air foraging species that collects plant litter and wood debris to cultivate fungi in fungal gardens [82], termites may act as vectors, transmitting deltaflexivirus to other fungi. Furthermore, the viruses we identified, typically associated with fungi, also deepen their connections with species from other kingdoms on the tanglegram tree. For example, while Partitiviridae are naturally associated with fungi and plants, NcPV also shows close connections with Metazoa. In fact, based largely on phylogenetic predictions, various eukaryotic viruses have been found to undergo horizontal transfer between organisms of plants, fungi, and animals [83]. The rice dwarf virus was demonstrated to infect both plant and insect vectors [84]; moreover, plant-infecting rhabdoviruses, tospoviruses, and tenuiviruses are now known to replicate and spread in vector insects and shuttle between plants and animals [85]. Furthermore, Bian et al. demonstrated that plant virus infection in plants enables Cryphonectria hypovirus 1 to undergo horizontal transfer from fungi to plants and other heterologous fungal species [86].
Recent studies have greatly expanded the diversity of mycoviruses [87, 88]. Gilbert et al. [20] investigated publicly available fungal transcriptomes from the subphylum Pezizomycotina, resulting in the detection of 52 novel mycoviruses; Myers et al. [18] employed both culture-based and transcriptome-mining approaches to identify 85 unique RNA viruses across 333 fungi; Ruiz-Padilla et al. identified 62 new mycoviral species from 248 Botrytis cinerea field isolates; Zhou et al. identified 20 novel viruses from 90 fungal strains (across four different macrofungi species) [89]. However, compared to these studies, our work identified fewer novel viruses, possibly due to the following reasons: 1) The libraries from the same Bioproject are usually from the same strains (or isolates). Therefore, there is a certain degree of redundancy in the datasets collected for this study. 2) Contigs shorter than 1,500 nt were discarded, potentially resulting in the oversight of short viral molecules. 3) Establishing a threshold of 70% aa sequence identity may also lead to the exclusion of certain viruses. 4) Some poly(A)-enriched RNA-seq libraries are likely to miss non-polyadenylated RNA viral genomes.
Taxonomy is a dynamic science, evolving with improvements in analytical methods and the emergence of new data. Identifying and rectifying incorrect classifications when new information becomes available is an ongoing and inevitable process in today's rapidly expanding field of virology. For instance, in 1975, members of the genera Rubivirus and Alphavirus were initially grouped under the family Togaviridae; however, in 2019, Rubivirus was reclassified into the family Matonaviridae due to recognized differences in transmission modes and virion structures [90]. Additionally, the conflicts between certain members of the genera Magoulivirus and Gammapartitivirus mentioned here and their current demarcation criteria (e.g., amino acid identity, nucleotide length thresholds) need to be reconsidered.
Taken together, these findings reveal the potential diversity and novelty within fungal-associated viral communities and discuss the genetic similarities among different fungal-associated viruses. These findings advance our understanding of fungal-associated viruses and suggest the importance of subsequent in-depth investigations into the interactions between fungi and viruses, which will shed light on the important roles of these viruses in the global fungal kingdom.
Availability of data and materials
The data reported in this paper have been deposited in the GenBase in National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences/China National Center for Bioinformation, under accession numbers C_AA066339.1-C_AA066350.1 that are publicly accessible at https://ngdc.cncb.ac.cn/genbase. Please refer to Table 1 for details.
References
Leigh DM, Peranic K, Prospero S, Cornejo C, Curkovic-Perica M, Kupper Q, et al. Long-read sequencing reveals the evolutionary drivers of intra-host diversity across natural RNA mycovirus infections. Virus Evol. 2021;7(2):veab101. https://doi.org/10.1093/ve/veab101. Epub 2022/03/19 PubMed PMID: 35299787; PubMed Central PMCID: PMCPMC8923234.
Ghabrial SA, Suzuki N. Viruses of plant pathogenic fungi. Annu Rev Phytopathol. 2009;47:353–84. https://doi.org/10.1146/annurev-phyto-080508-081932. Epub 2009/04/30 PubMed PMID: 19400634.
Ghabrial SA, Caston JR, Jiang D, Nibert ML, Suzuki N. 50-plus years of fungal viruses. Virology. 2015;479–480:356–68. https://doi.org/10.1016/j.virol.2015.02.034. Epub 2015/03/17 PubMed PMID: 25771805.
Chen YM, Sadiq S, Tian JH, Chen X, Lin XD, Shen JJ, et al. RNA viromes from terrestrial sites across China expand environmental viral diversity. Nat Microbiol. 2022;7(8):1312–23. https://doi.org/10.1038/s41564-022-01180-2. Epub 2022/07/29 PubMed PMID: 35902778.
Pearson MN, Beever RE, Boine B, Arthur K. Mycoviruses of filamentous fungi and their relevance to plant pathology. Mol Plant Pathol. 2009;10(1):115–28. https://doi.org/10.1111/j.1364-3703.2008.00503.x. Epub 2009/01/24 PubMed PMID: 19161358; PubMed Central PMCID: PMCPMC6640375.
Santiago-Rodriguez TM, Hollister EB. Unraveling the viral dark matter through viral metagenomics. Front Immunol. 2022;13:1005107. https://doi.org/10.3389/fimmu.2022.1005107. Epub 2022/10/04 PubMed PMID: 36189246; PubMed Central PMCID: PMCPMC9523745.
Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol. 2008;159(5):349–57. https://doi.org/10.1016/j.resmic.2008.04.010. Epub 2008/06/21 PubMed PMID: 18565737.
Guo W, Yan H, Ren X, Tang R, Sun Y, Wang Y, et al. Berberine induces resistance against tobacco mosaic virus in tobacco. Pest Manag Sci. 2020;76(5):1804–13. https://doi.org/10.1002/ps.5709. Epub 2019/12/10 PubMed PMID: 31814252.
Villabruna N, Izquierdo-Lara RW, Schapendonk CME, de Bruin E, Chandler F, Thao TTN, et al. Profiling of humoral immune responses to norovirus in children across Europe. Sci Rep. 2022;12(1):14275. https://doi.org/10.1038/s41598-022-18383-6. Epub 2022/08/23 PubMed PMID: 35995986.
Zhang Y, Gao J, Li Y. Diversity of mycoviruses in edible fungi. Virus Genes. 2022;58(5):377–91. https://doi.org/10.1007/s11262-022-01908-6. Epub 2022/06/07 PubMed PMID: 35668282.
Shkoporov AN, Clooney AG, Sutton TDS, Ryan FJ, Daly KM, Nolan JA, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26(4):527–41. https://doi.org/10.1016/j.chom.2019.09.009. Epub 2019/10/11 PubMed PMID: 31600503.
Botella L, Janousek J, Maia C, Jung MH, Raco M, Jung T. Marine Oomycetes of the Genus Halophytophthora harbor viruses related to Bunyaviruses. Front Microbiol. 2020;11:1467. https://doi.org/10.3389/fmicb.2020.01467. Epub 2020/08/08 PubMed PMID: 32760358; PubMed Central PMCID: PMCPMC7375090.
Kotta-Loizou I. Mycoviruses and their role in fungal pathogenesis. Curr Opin Microbiol. 2021;63:10–8. https://doi.org/10.1016/j.mib.2021.05.007. Epub 2021/06/09 PubMed PMID: 34102567.
Ellis LF, Kleinschmidt WJ. Virus-like particles of a fraction of statolon, a mould product. Nature. 1967;215(5101):649–50. https://doi.org/10.1038/215649a0. Epub 1967/08/05 PubMed PMID: 6050227.
Banks GT, Buck KW, Chain EB, Himmelweit F, Marks JE, Tyler JM, et al. Viruses in fungi and interferon stimulation. Nature. 1968;218(5141):542–5. https://doi.org/10.1038/218542a0. Epub 1968/05/11 PubMed PMID: 4967851.
Jia J, Fu Y, Jiang D, Mu F, Cheng J, Lin Y, et al. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol. 2021;7(1):veab032. https://doi.org/10.1093/ve/veab032.
Mu F, Li B, Cheng S, Jia J, Jiang D, Fu Y, et al. Nine viruses from eight lineages exhibiting new evolutionary modes that co-infect a hypovirulent phytopathogenic fungus. Plos Pathog. 2021;17(8):e1009823. https://doi.org/10.1371/journal.ppat.1009823. Epub 2021/08/25 PubMed PMID: 34428260; PubMed Central PMCID: PMCPMC8415603.
Myers JM, Bonds AE, Clemons RA, Thapa NA, Simmons DR, Carter-House D, et al. Survey of early-diverging lineages of fungi reveals abundant and diverse Mycoviruses. mBio. 2020;11(5):e02027. https://doi.org/10.1128/mBio.02027-20. Epub 2020/09/10 PubMed PMID: 32900807; PubMed Central PMCID: PMCPMC7482067.
Ruiz-Padilla A, Rodriguez-Romero J, Gomez-Cid I, Pacifico D, Ayllon MA. Novel Mycoviruses discovered in the Mycovirome of a Necrotrophic fungus. MBio. 2021;12(3):e03705. https://doi.org/10.1128/mBio.03705-20. Epub 2021/05/13 PubMed PMID: 33975945; PubMed Central PMCID: PMCPMC8262958.
Gilbert KB, Holcomb EE, Allscheid RL, Carrington JC. Hiding in plain sight: new virus genomes discovered via a systematic analysis of fungal public transcriptomes. Plos One. 2019;14(7):e0219207. https://doi.org/10.1371/journal.pone.0219207. Epub 2019/07/25 PubMed PMID: 31339899; PubMed Central PMCID: PMCPMC6655640.
Khan HA, Telengech P, Kondo H, Bhatti MF, Suzuki N. Mycovirus hunting revealed the presence of diverse viruses in a single isolate of the Phytopathogenic fungus diplodia seriata from Pakistan. Front Cell Infect Microbiol. 2022;12:913619. https://doi.org/10.3389/fcimb.2022.913619. Epub 2022/07/19 PubMed PMID: 35846770; PubMed Central PMCID: PMCPMC9277117.
Kotta-Loizou I, Coutts RHA. Mycoviruses in Aspergilli: a comprehensive review. Front Microbiol. 2017;8:1699. https://doi.org/10.3389/fmicb.2017.01699. Epub 2017/09/22 PubMed PMID: 28932216; PubMed Central PMCID: PMCPMC5592211.
Garcia-Pedrajas MD, Canizares MC, Sarmiento-Villamil JL, Jacquat AG, Dambolena JS. Mycoviruses in biological control: from basic research to field implementation. Phytopathology. 2019;109(11):1828–39. https://doi.org/10.1094/PHYTO-05-19-0166-RVW. Epub 2019/08/10 PubMed PMID: 31398087.
Rigling D, Prospero S. Cryphonectria parasitica, the causal agent of chestnut blight: invasion history, population biology and disease control. Mol Plant Pathol. 2018;19(1):7–20. https://doi.org/10.1111/mpp.12542. Epub 2017/02/01 PubMed PMID: 28142223; PubMed Central PMCID: PMCPMC6638123.
Okada R, Ichinose S, Takeshita K, Urayama SI, Fukuhara T, Komatsu K, et al. Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: down-regulation of host growth and up-regulation of host plant pathogenicity. Virology. 2018;519:23–32. https://doi.org/10.1016/j.virol.2018.03.027. Epub 2018/04/10 PubMed PMID: 29631173.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. https://doi.org/10.1038/nmeth.1923. Epub 2012/03/06 PubMed PMID: 22388286; PubMed Central PMCID: PMCPMC3322381.
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes De Novo assembler. Curr Protoc Bioinform. 2020;70(1):e102. https://doi.org/10.1002/cpbi.102. Epub 2020/06/20 PubMed PMID: 32559359.
Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. https://doi.org/10.1016/j.ymeth.2016.02.020. Epub 2016/03/26 PubMed PMID: 27012178.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–95. https://doi.org/10.1093/bioinformatics/btp698. Epub 2010/01/19 PubMed PMID: 20080505; PubMed Central PMCID: PMCPMC2828108.
Roux S, Paez-Espino D, Chen IA, Palaniappan K, Ratner A, Chu K, et al. IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021;49(D1):D764–75. https://doi.org/10.1093/nar/gkaa946. Epub 2020/11/03 PubMed PMID: 33137183; PubMed Central PMCID: PMCPMC7778971.
Mirdita M, Steinegger M, Soding J. MMseqs2 desktop and local web server app for fast, interactive sequence searches. Bioinformatics. 2019;35(16):2856–8. https://doi.org/10.1093/bioinformatics/bty1057. Epub 2019/01/08 PubMed PMID: 30615063; PubMed Central PMCID: PMCPMC6691333.
Buchfink B, Reuter K, Drost HG. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods. 2021;18(4):366–8. https://doi.org/10.1038/s41592-021-01101-x. Epub 2021/04/09 PubMed PMID: 33828273; PubMed Central PMCID: PMCPMC8026399.
Shen W, Ren H. TaxonKit: A practical and efficient NCBI taxonomy toolkit. J Genet Genomics. 2021;48(9):844–50. https://doi.org/10.1016/j.jgg.2021.03.006. Epub 2021/05/19 PubMed PMID: 34001434.
Gautam A, Felderhoff H, Bagci C, Huson DH. Using AnnoTree to get more assignments, faster, in DIAMOND+MEGAN microbiome analysis. mSystems. 2022;7(1):e0140821. https://doi.org/10.1128/msystems.01408-21. Epub 2022/02/23 PubMed PMID: 35191776; PubMed Central PMCID: PMCPMC8862659.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. https://doi.org/10.1093/molbev/msy096. Epub 2018/05/04 PubMed PMID: 29722887; PubMed Central PMCID: PMCPMC5967553.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4. https://doi.org/10.1093/molbev/msaa015. Epub 2020/02/06 PubMed PMID: 32011700; PubMed Central PMCID: PMCPMC7182206.
Letunic I, Bork P. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res (2024). https://doi.org/10.1093/nar/gkae268
Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. Plos One. 2014;9(9):e108277. https://doi.org/10.1371/journal.pone.0108277. Epub 2014/09/27 PubMed PMID: 25259891; PubMed Central PMCID: PMCPMC4178126.
Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, et al. TimeTree 5: an expanded resource for species divergence times. Mol Biol Evol. 2022;39(8):msac174. https://doi.org/10.1093/molbev/msac174. Epub 2022/08/07 PubMed PMID: 35932227; PubMed Central PMCID: PMCPMC9400175.
Revell LJ. phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things). PeerJ. 2024;12:e16505. https://doi.org/10.7717/peerj.16505. Epub 2024/01/09 PubMed PMID: 38192598; PubMed Central PMCID: PMCPMC10773453.
Santichaivekin S, Yang Q, Liu J, Mawhorter R, Jiang J, Wesley T, et al. eMPRess: a systematic cophylogeny reconciliation tool. Bioinformatics. 2021;37(16):2481–2. https://doi.org/10.1093/bioinformatics/btaa978. Epub 2020/11/21 PubMed PMID: 33216126.
Ma W, Smirnov D, Libeskind-Hadas R. DTL reconciliation repair. BMC Bioinformatics. 2017;18(Suppl 3):76. https://doi.org/10.1186/s12859-017-1463-9. Epub 2017/04/01 PubMed PMID: 28361686; PubMed Central PMCID: PMCPMC5374596.
Members C-N, Partners. Database resources of the national genomics data center, China national center for bioinformation in 2024. Nucleic Acids Res. 2024;52(D1):D18–32. https://doi.org/10.1093/nar/gkad1078. Epub 2023/11/29 PubMed PMID: 38018256; PubMed Central PMCID: PMCPMC10767964.
Shafik K, Umer M, You H, Aboushedida H, Wang Z, Ni D, et al. Characterization of a Novel Mitovirus infecting Melanconiella theae isolated from tea plants. Front Microbiol. 2021;12: 757556. https://doi.org/10.3389/fmicb.2021.757556. Epub 2021/12/07 PubMed PMID: 34867881; PubMed Central PMCID: PMCPMC8635788
Kamaruzzaman M, He G, Wu M, Zhang J, Yang L, Chen W, et al. A novel Partitivirus in the Hypovirulent isolate QT5–19 of the plant pathogenic fungus Botrytis cinerea. Viruses. 2019;11(1):24. https://doi.org/10.3390/v11010024. Epub 2019/01/06 PubMed PMID: 30609795; PubMed Central PMCID: PMCPMC6356794.
Akata I, Keskin E, Sahin E. Molecular characterization of a new mitovirus hosted by the ectomycorrhizal fungus Albatrellopsis flettii. Arch Virol. 2021;166(12):3449–54. https://doi.org/10.1007/s00705-021-05250-4. Epub 2021/09/24 PubMed PMID: 34554305.
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, et al. Recent changes to virus taxonomy ratified by the international committee on taxonomy of viruses (2022). Arch Virol. 2022;167(11):2429–40. https://doi.org/10.1007/s00705-022-05516-5. Epub 2022/08/24 PubMed PMID: 35999326; PubMed Central PMCID: PMCPMC10088433.
Alvarez-Quinto R, Grinstead S, Jones R, Mollov D. Complete genome sequence of a new mitovirus associated with walking iris (Trimezia northiana). Arch Virol. 2023;168(11):273. https://doi.org/10.1007/s00705-023-05901-8. Epub 2023/10/17 PubMed PMID: 37845386.
Gilmer D, Ratti C, Ictv RC. ICTV Virus taxonomy profile: Benyviridae. J Gen Virol. 2017;98(7):1571–2. https://doi.org/10.1099/jgv.0.000864. Epub 2017/07/18 PubMed PMID: 28714846; PubMed Central PMCID: PMCPMC5656776.
Wetzel V, Willlems G, Darracq A, Galein Y, Liebe S, Varrelmann M. The Beta vulgaris-derived resistance gene Rz2 confers broad-spectrum resistance against soilborne sugar beet-infecting viruses from different families by recognizing triple gene block protein 1. Mol Plant Pathol. 2021;22(7):829–42. https://doi.org/10.1111/mpp.13066. Epub 2021/05/06 PubMed PMID: 33951264; PubMed Central PMCID: PMCPMC8232027.
Ayllon MA, Turina M, Xie J, Nerva L, Marzano SL, Donaire L, et al. ICTV Virus taxonomy profile: Botourmiaviridae. J Gen Virol. 2020;101(5):454–5. https://doi.org/10.1099/jgv.0.001409. Epub 2020/05/08 PubMed PMID: 32375992; PubMed Central PMCID: PMCPMC7414452.
Xiao J, Wang X, Zheng Z, Wu Y, Wang Z, Li H, et al. Molecular characterization of a novel deltaflexivirus infecting the edible fungus Pleurotus ostreatus. Arch Virol. 2023;168(6):162. https://doi.org/10.1007/s00705-023-05789-4. Epub 2023/05/17 PubMed PMID: 37195309.
Canuti M, Rodrigues B, Lang AS, Dufour SC, Verhoeven JTP. Novel divergent members of the Kitrinoviricota discovered through metagenomics in the intestinal contents of red-backed voles (Clethrionomys gapperi). Int J Mol Sci. 2022;24(1):131. https://doi.org/10.3390/ijms24010131. Epub 2023/01/09 PubMed PMID: 36613573; PubMed Central PMCID: PMCPMC9820622.
Hermanns K, Zirkel F, Kopp A, Marklewitz M, Rwego IB, Estrada A, et al. Discovery of a novel alphavirus related to Eilat virus. J Gen Virol. 2017;98(1):43–9. https://doi.org/10.1099/jgv.0.000694. Epub 2017/02/17 PubMed PMID: 28206905.
Jiang D, Ayllon MA, Marzano SL, Ictv RC. ICTV Virus taxonomy profile: Mymonaviridae. J Gen Virol. 2019;100(10):1343–4. https://doi.org/10.1099/jgv.0.001301. Epub 2019/09/04 PubMed PMID: 31478828.
Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, et al. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci U S A. 2014;111(33):12205–10. https://doi.org/10.1073/pnas.1401786111. Epub 2014/08/06 PubMed PMID: 25092337; PubMed Central PMCID: PMCPMC4143027.
Zhong J, Li P, Gao BD, Zhong SY, Li XG, Hu Z, et al. Novel and diverse mycoviruses co-infecting a single strain of the phytopathogenic fungus Alternaria dianthicola. Front Cell Infect Microbiol. 2022;12:980970. https://doi.org/10.3389/fcimb.2022.980970. Epub 2022/10/15 PubMed PMID: 36237429; PubMed Central PMCID: PMCPMC9552818.
Wang W, Wang X, Tu C, Yang M, Xiang J, Wang L, et al. Novel Mycoviruses discovered from a Metatranscriptomics survey of the Phytopathogenic Alternaria Fungus. Viruses. 2022;14(11):2552. https://doi.org/10.3390/v14112552. Epub 2022/11/25 PubMed PMID: 36423161; PubMed Central PMCID: PMCPMC9693364.
Sun Y, Li J, Gao GF, Tien P, Liu W. Bunyavirales ribonucleoproteins: the viral replication and transcription machinery. Crit Rev Microbiol. 2018;44(5):522–40. https://doi.org/10.1080/1040841X.2018.1446901. Epub 2018/03/09 PubMed PMID: 29516765.
Li P, Bhattacharjee P, Gagkaeva T, Wang S, Guo L. A novel bipartite negative-stranded RNA mycovirus of the order Bunyavirales isolated from the phytopathogenic fungus Fusarium sibiricum. Arch Virol. 2023;169(1):13. https://doi.org/10.1007/s00705-023-05942-z. Epub 2023/12/29 PubMed PMID: 38155262.
Ferron F, Weber F, de la Torre JC, Reguera J. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res. 2017;234:118–34. https://doi.org/10.1016/j.virusres.2017.01.018. Epub 2017/02/01 PubMed PMID: 28137457; PubMed Central PMCID: PMCPMC7114536.
Vainio EJ, Chiba S, Ghabrial SA, Maiss E, Roossinck M, Sabanadzovic S, et al. ICTV Virus taxonomy profile: Partitiviridae. J Gen Virol. 2018;99(1):17–8. https://doi.org/10.1099/jgv.0.000985. Epub 2017/12/08 PubMed PMID: 29214972; PubMed Central PMCID: PMCPMC5882087.
Mifsud JCO, Gallagher RV, Holmes EC, Geoghegan JL. Transcriptome mining expands knowledge of RNA viruses across the plant Kingdom. J Virol. 2022;96(24):e0026022. https://doi.org/10.1128/jvi.00260-22. Epub 2022/06/01 PubMed PMID: 35638822; PubMed Central PMCID: PMCPMC9769393.
Tamada T, Kondo H. Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors. J Gen Plant Pathol. 2013;79:307–20.
Dolja VV, Krupovic M, Koonin EV. Deep roots and splendid boughs of the global plant virome. Annu Rev Phytopathol. 2020;58:23–53.
Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, et al. Global organization and proposed Megataxonomy of the virus world. Microbiol Mol Biol Rev. 2020;84(2):e00061. https://doi.org/10.1128/MMBR.00061-19. Epub 2020/03/07 PubMed PMID: 32132243; PubMed Central PMCID: PMCPMC7062200.
Osono T. Role of phyllosphere fungi of forest trees in the development of decomposer fungal communities and decomposition processes of leaf litter. Can J Microbiol. 2006;52(8):701–16. https://doi.org/10.1139/w06-023. Epub 2006/08/19 PubMed PMID: 16917528.
Li Z, Pan D, Wei G, Pi W, Zhang C, Wang JH, et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J. 2021;15(8):2366–78. https://doi.org/10.1038/s41396-021-00932-y. Epub 2021/03/03 PubMed PMID: 33649554; PubMed Central PMCID: PMCPMC8319345.
Hierweger MM, Koch MC, Rupp M, Maes P, Di Paola N, Bruggmann R, et al. Novel Filoviruses, Hantavirus, and Rhabdovirus in freshwater fish, Switzerland, 2017. Emerg Infect Dis. 2021;27(12):3082–91. https://doi.org/10.3201/eid2712.210491. Epub 2021/11/23 PubMed PMID: 34808081; PubMed Central PMCID: PMCPMC8632185.
La Rosa G, Iaconelli M, Mancini P, Bonanno Ferraro G, Veneri C, Bonadonna L, et al. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci Total Environ. 2020;736:139652. https://doi.org/10.1016/j.scitotenv.2020.139652. Epub 2020/05/29 PubMed PMID: 32464333; PubMed Central PMCID: PMCPMC7245320.
Sutela S, Poimala A, Vainio EJ. Viruses of fungi and oomycetes in the soil environment. FEMS Microbiol Ecol. 2019;95(9):fiz119. https://doi.org/10.1093/femsec/fiz119. Epub 2019/08/01 PubMed PMID: 31365065.
Yu X, Li B, Fu Y, Jiang D, Ghabrial SA, Li G, et al. A geminivirus-related DNA mycovirus that confers hypovirulence to a plant pathogenic fungus. Proc Natl Acad Sci U S A. 2010;107(18):8387–92. https://doi.org/10.1073/pnas.0913535107. Epub 2010/04/21 PubMed PMID: 20404139; PubMed Central PMCID: PMCPMC2889581.
Li P, Wang S, Zhang L, Qiu D, Zhou X, Guo L. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci Adv. 2020;6(14):eaay9634. https://doi.org/10.1126/sciadv.aay9634. Epub 2020/04/15 PubMed PMID: 32284975; PubMed Central PMCID: PMCPMC7138691.
Khalifa ME, MacDiarmid RM. A mechanically transmitted DNA Mycovirus is targeted by the defence machinery of its host, Botrytis cinerea. Viruses. 2021;13(7):1315. https://doi.org/10.3390/v13071315. Epub 2021/08/11 PubMed PMID: 34372522; PubMed Central PMCID: PMCPMC8309985.
Yu X, Li B, Fu Y, Xie J, Cheng J, Ghabrial SA, et al. Extracellular transmission of a DNA mycovirus and its use as a natural fungicide. Proc Natl Acad Sci U S A. 2013;110(4):1452–7. https://doi.org/10.1073/pnas.1213755110. Epub 2013/01/09 PubMed PMID: 23297222; PubMed Central PMCID: PMCPMC3557086.
Nuss DL. Hypovirulence: mycoviruses at the fungal-plant interface. Nat Rev Microbiol. 2005;3(8):632–42. https://doi.org/10.1038/nrmicro1206. Epub 2005/08/03 PubMed PMID: 16064055.
Coenen A, Kevei F, Hoekstra RF. Factors affecting the spread of double-stranded RNA viruses in Aspergillus nidulans. Genet Res. 1997;69(1):1–10. https://doi.org/10.1017/s001667239600256x. Epub 1997/02/01 PubMed PMID: 9164170.
Freitas CSA, Maciel LF, Dos Correa Santos RA, Costa O, Maia FCB, Rabelo RS, et al. Bacterial volatile organic compounds induce adverse ultrastructural changes and DNA damage to the sugarcane pathogenic fungus Thielaviopsis ethacetica. Environ Microbiol. 2022;24(3):1430–53. https://doi.org/10.1111/1462-2920.15876. Epub 2022/01/08 PubMed PMID: 34995419.
Martinez-Alvarez P, Vainio EJ, Botella L, Hantula J, Diez JJ. Three mitovirus strains infecting a single isolate of Fusarium circinatum are the first putative members of the family Narnaviridae detected in a fungus of the genus Fusarium. Arch Virol. 2014;159(8):2153–5. https://doi.org/10.1007/s00705-014-2012-8. Epub 2014/02/13 PubMed PMID: 24519462.
Deakin G, Dobbs E, Bennett JM, Jones IM, Grogan HM, Burton KS. Multiple viral infections in Agaricus bisporus - characterisation of 18 unique RNA viruses and 8 ORFans identified by deep sequencing. Sci Rep. 2017;7(1):2469. https://doi.org/10.1038/s41598-017-01592-9. Epub 2017/05/28 PubMed PMID: 28550284; PubMed Central PMCID: PMCPMC5446422.
Litov AG, Zueva AI, Tiunov AV, Van Thinh N, Belyaeva NV, Karganova GG. Virome of three termite species from Southern Vietnam. Viruses. 2022;14(5):860. https://doi.org/10.3390/v14050860. Epub 2022/05/29 PubMed PMID: 35632601; PubMed Central PMCID: PMCPMC9143207.
Hu J, Neoh KB, Appel AG, Lee CY. Subterranean termite open-air foraging and tolerance to desiccation: Comparative water relation of two sympatric Macrotermes spp. (Blattodea: Termitidae). Comp Biochem Physiol A Mol Integr Physiol. 2012;161(2):201–7. https://doi.org/10.1016/j.cbpa.2011.10.028. Epub 2011/11/17 PubMed PMID: 22085890.
Kondo H, Botella L, Suzuki N. Mycovirus diversity and evolution revealed/inferred from recent studies. Annu Rev Phytopathol. 2022;60:307–36. https://doi.org/10.1146/annurev-phyto-021621-122122. Epub 2022/05/25 PubMed PMID: 35609970.
Fukushi T. Relationships between propagative rice viruses and their vectors. 1969.
Sun L, Kondo H, Bagus AI. Cross-kingdom virus infection. Encyclopedia of Virology: Volume 1–5. 4th Ed. Elsevier; 2020. pp. 443–9. https://doi.org/10.1016/B978-0-12-809633-8.21320-4.
Bian R, Andika IB, Pang T, Lian Z, Wei S, Niu E, et al. Facilitative and synergistic interactions between fungal and plant viruses. Proc Natl Acad Sci U S A. 2020;117(7):3779–88. https://doi.org/10.1073/pnas.1915996117. Epub 2020/02/06 PubMed PMID: 32015104; PubMed Central PMCID: PMCPMC7035501.
Chiapello M, Rodriguez-Romero J, Ayllon MA, Turina M. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol. 2020;6(2):veaa058. https://doi.org/10.1093/ve/veaa058. Epub 2020/12/17 PubMed PMID: 33324489; PubMed Central PMCID: PMCPMC7724247.
Sutela S, Forgia M, Vainio EJ, Chiapello M, Daghino S, Vallino M, et al. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol. 2020;6(2):veaa076. https://doi.org/10.1093/ve/veaa076. Epub 2020/12/17 PubMed PMID: 33324490; PubMed Central PMCID: PMCPMC7724248.
Zhou K, Zhang F, Deng Y. Comparative analysis of viromes identified in multiple macrofungi. Viruses. 2024;16(4):597. https://doi.org/10.3390/v16040597. Epub 2024/04/27 PubMed PMID: 38675938; PubMed Central PMCID: PMCPMC11054281.
Siddell SG, Smith DB, Adriaenssens E, Alfenas-Zerbini P, Dutilh BE, Garcia ML, et al. Virus taxonomy and the role of the International Committee on Taxonomy of Viruses (ICTV). J Gen Virol. 2023;104(5):001840. https://doi.org/10.1099/jgv.0.001840. Epub 2023/05/04 PubMed PMID: 37141106; PubMed Central PMCID: PMCPMC10227694.
Acknowledgements
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; WZ and CZ contributed to the conception and design; XL, ZD, JXU, WL and PN contributed to the collection and assembly of data; XL, ZD and JXE contributed to the data analysis and interpretation.
Funding
This research was supported by National Key Research and Development Programs of China [No.2023YFD1801301 and 2022YFC2603801] and the National Natural Science Foundation of China [No.82341106].
Author information
Authors and Affiliations
Contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript; WZ and CZ contributed to the conception and design; XL, ZD, JXU, WL and PN contributed to the collection and assembly of data; XL, ZD and JXE contributed to the data analysis and interpretation.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lu, X., Dai, Z., Xue, J. et al. Discovery of novel RNA viruses through analysis of fungi-associated next-generation sequencing data. BMC Genomics 25, 517 (2024). https://doi.org/10.1186/s12864-024-10432-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10432-w