- Research article
- Open access
- Published:
Whole genome de novo sequencing and comparative genomic analyses suggests that Chlamydia psittaci strain 84/2334 should be reclassified as Chlamydia abortus species
BMC Genomics volume 22, Article number: 159 (2021)
Abstract
Background
Chlamydia abortus and Chlamydia psittaci are important pathogens of livestock and avian species, respectively. While C. abortus is recognized as descended from C. psittaci species, there is emerging evidence of strains that are intermediary between the two species, suggesting they are recent evolutionary ancestors of C. abortus. Such strains include C. psittaci strain 84/2334 that was isolated from a parrot. Our aim was to classify this strain by sequencing its genome and explore its evolutionary relationship to both C. abortus and C. psittaci.
Results
In this study, methods based on multi-locus sequence typing (MLST) of seven housekeeping genes and on typing of five species discriminant proteins showed that strain 84/2334 clustered with C. abortus species. Furthermore, whole genome de novo sequencing of the strain revealed greater similarity to C. abortus in terms of GC content, while 16S rRNA and whole genome phylogenetic analysis, as well as network and recombination analysis showed that the strain clusters more closely with C. abortus strains. The analysis also suggested a closer evolutionary relationship between this strain and the major C. abortus clade, than to two other intermediary avian C. abortus strains or C. psittaci strains. Molecular analyses of genes (polymorphic membrane protein and transmembrane head protein genes) and loci (plasticity zone), found in key virulence-associated regions that exhibit greatest diversity within and between chlamydial species, reveal greater diversity than present in sequenced C. abortus genomes as well as similar features to both C. abortus and C. psittaci species. The strain also possesses an extrachromosomal plasmid, as found in most C. psittaci species but absent from all sequenced classical C. abortus strains.
Conclusion
Overall, the results show that C. psittaci strain 84/2334 clusters very closely with C. abortus strains, and are consistent with the strain being a recent C. abortus ancestral species. This suggests that the strain should be reclassified as C. abortus. Furthermore, the identification of a C. abortus strain bearing an extra-chromosomal plasmid has implications for plasmid-based transformation studies to investigate gene function as well as providing a potential route for the development of a next generation vaccine to protect livestock from C. abortus infection.
Introduction
The family Chlamydiaceae comprises a group of obligate intracellular Gram-negative bacteria that are responsible for a broad range of infections in non-human mammals, birds and humans [1]. Since 1999 the family has undergone a number of taxonomic reclassifications at the genus and species levels based on sequence analysis of the 16S and 23S rRNA genes, as well as taking into account biological differences [2,3,4,5]. Current classification within this family recognises a single genus, Chlamydia, and 14 species (Chlamydia abortus, Chlamydia avium, Chlamydia buteonis, Chlamydia caviae, Chlamydia felis, Chlamydia gallinacea, Chlamydia muridarum, Chlamydia pecorum, Chlamydia pneumoniae, Chlamydia poikilothermis, Chlamydia psittaci, Chlamydia serpentis, Chlamydia suis and Chlamydia trachomatis) [6,7,8], plus a further four Candidatus species (Ca. Chlamydia ibidis, Ca. Chlamydia corallus, Ca. Chlamydia sanzinia and Ca. Chlamydia testudinis) [9,10,11,12].
One of the oldest recognised chlamydial species is C. psittaci that is associated with respiratory, ocular and enteric infections in psittacine birds and poultry [1, 13]. Shedding of the pathogen in faecal, respiratory and ocular secretions can occur and result in zoonotic infections in humans, leading to pneumonia (psittacosis) that can be fatal [13, 14]. Such infections have been frequently reported in veterinarians, breeders, pet shop and poultry workers [1, 14]. C. psittaci has been classified into nine genotypes based on ompA (major outer membrane protein gene) sequencing [15], of which seven (A-F, E/B) are each generally associated with specific species of birds, while a further two were isolated from mammalian species, specifically cattle (WC) and muskrat (M56) [16]. In addition, a further ten provisional genotypes (1V, Mat116, YP84, R54, 6N, CPX0308, I, J, G, G1 and G2) have been described [17,18,19]. More recently, three newly described species, C. gallinacea, C. avium and C. buteonis, have also been found in birds [11, 20], with C. buteonis suggested to have a phylogenetic intermediate position between C. psittaci and C. abortus species [7].
Chlamydial infections in non-avian livestock species are principally caused by three species, C. abortus, C. pecorum and C. suis [1], of which C. abortus is considered the most closely related to and suggested to be evolved from C. psittaci species [21]. C. abortus is principally responsible for causing enzootic abortion (syn. ovine enzootic abortion, OEA; enzootic abortion of ewes, EAE; ovine chlamydiosis) in sheep and goats [1, 22]. This organism, which was first described in 1950 [23], generally causes abortion in the last 2 to 3 weeks of pregnancy or the birth of weak or stillborn lambs or kids. C. abortus also causes sporadic reproductive failure in cattle, horses and pigs and the bacterium presents a dangerous zoonotic risk to pregnant women, in whom it can cause spontaneous abortion, as well as being potentially fatal for the woman [24,25,26]. In contrast to C. psittaci, the genome of C. abortus is considered to be relatively stable with very little diversity and undergoing little or no recombination [27].
It has long been recognised that there are a number of C. psittaci strains that differ from the classical avian C. psittaci strains, with comparative sequence and MLST analysis suggesting that they are more closely related to C. abortus species [21, 28]. Such strains have been classified as belonging to genotype F by ompA AluI typing, and include isolates Prk/Daruma (isolated from parakeets) [29], 10,433-MA (isolated from a parrot) [30], VS225 (isolated from a parakeet) [2], and 84/2334 (isolated from a yellow-crowned Amazon parrot) [31]. These isolates, as well as others recently described with provisional genotypes 1V, G1 and G2 [18], have been variously referred to as atypical C. psittaci/C. abortus, C. psittaci/C.abortus intermediate and avian C. abortus strains [21]. The purpose of this study was to characterise and sequence the genome of one of these atypical C. psittaci strains, specifically strain 84/2334, and determine its similarity and genetic relatedness to both C. psittaci and C. abortus species through comparative sequence analysis.
Results
Molecular typing of Chlamydia psittaci strain 84/2334
PhyML phylogenetic analysis of a 3147 bp alignment of concatenated fragments of seven multi-locus sequence typing (MLST) housekeeping genes (enoA, fumC, gatA, gidA, hemN, hlfX and oppA) [28, 32] for C. psittaci strain 84/2334 as well as other representative strains from each of the currently recognised Chlamydiaceae species are shown in Fig. 1. Strain 84/2334 was determined to cluster closely with the classical C. abortus strains, as previously observed using a less robust Neighbour-Joining method [28], as well as the three atypical avian C. abortus strains (15-70d/24, 15-49d/3 and 15-58d/44). Indeed, the strain appears more closely related to the classical C. abortus strains than to the avian C. abortus or C. psittaci strains, which is further supported by in silico genome-to-genome distance comparison [33] with published reference C. abortus and C. psittaci genomes (Additional file 1). In addition, it was noted that C. buteonis strain RSHA appears to cluster with C. psittaci species, but having a phylogenetic position somewhere between C. psittaci and C. abortus, as previously reported [7]. Cluster analysis of the allelic profiles or sequence type (ST) of each strain in minimum spanning trees produced a similar result (Additional file 2), with strain 84/2334 clustering with the C. abortus strains. Phylogenetic analysis of the 16S rRNA sequences also produced a similar tree structure to that obtained by MLST analysis (Additional file 3), the only exception being C. buteonis strain RSHA which clusters with C. abortus species, while ribosomal MLST (rMLST) based on 53 genes encoding the bacterial ribosome protein subunits (rps genes) [34] predicted the species taxon of strain 84/2334 as C. abortus with 100% support (Additional file 4).
MLST phylogenetic tree for C. psittaci strain 84/2334 and representative strains of Chlamydiaceae species. The consensus PhyML phylogenetic tree of a 3147 bp alignment of concatenated sequences of seven MLST housekeeping gene fragments (enoA, fumC, gatA, gidA, hemN, hlfX and oppA) was estimated in TOPALi by Maximum Likelihood using a GTR + G + I substitution and rate heterogeneity model, according to BIC model selection, and 100 non-parametric bootstrap replicates. The tree is rooted on the C. trachomatis/muridarum/suis branch and bootstrap support is indicated by the number at the node. The scale bar indicates the expected substitutions per site. Genotypes are given in square brackets. The tree was prepared for publication in Dendroscope. Strain 84/2334 is in bold and red font. Classical and avian C. abortus strains are in blue and green fonts, respectively
Utilising another typing system developed by Pillonel et al. [35] the protein sequences of five highly divergent proteins (Adk, FtsK, HemL, PepF and RpoN) used for distinguishing species were extracted for strain 84/2334 from the MLST database [36]. These sequences were compared to those of representative C. abortus (S26/3, AB7, LLG, 15-70d/24, 15-49d/3 and 15-58d/44) and C. psittaci (6BC, CP3, GR9, NJ1, MN, VS225, WS/RT/E30, M56 and WC) species and genotypes to determine the percentage sequence identity. Comparison of these sequence similarities for all five discriminant proteins with the threshold scores for distinguishing strains at the species level suggest that strain 84/2334 should be classified as C. abortus (Table 1).
Genomic sequencing of strain 84/2334
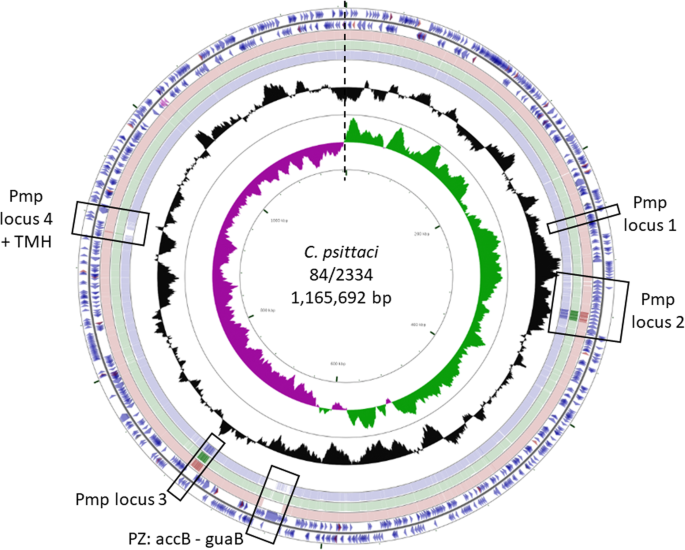
Genome sequencing and de novo assembly of C. psittaci strain 84/2334 successfully resulted in a single chromosomal contig with a 273x sequence coverage. The general features of the assembled genome in comparison to reference strains from C. abortus and C. psittaci species are shown in Table 2. The genome of C. psittaci strain 84/2334 comprises a single circular chromosome of 1,165,692 bp (Fig. 2) and in common with all other sequenced C. psittaci and C. abortus strains, 84/2334 has a single rRNA operon, 38 tRNA genes corresponding to all the amino acids except selenocysteine and pyrrolysine and a similar number of predicted coding sequences. Of note, the GC content of strain 84/2334 is 39.9%, which is similar to that of the C. abortus strains (Table 2).
Circular representation of the genome of C. psittaci strain 84/2334. Circles from the outside in show: the positions of protein-coding genes (blue), tRNA genes (orange) and rRNA genes (Pink) on the positive (circle 1) and negative (circle 2) strands, respectively. Circles 3–5 shows positions of BLAST hits determined through blastn comparisons of C. abortus S26/3 (circle 3), C. psittaci 6BC (circle 4) and C. psittaci GR9 (circle 5) with the following settings: query split size = 50,000 bp, query split overlap size = 0, expect value cut off = 0.00001. Low complexity sequences were eliminated from the analysis. The height of the shading in the BLAST results rings is proportional to the percent identity of the hit. Overlapping hits appear as darker shading. Circles 6 and 7 show plots of the GC content and GC skew plotted as the deviation from the average for the entire sequence. The origin of replication is indicated by the vertical dashed line. The four Pmp loci, PZ region and TMH loci are highlighted using rectangular boxes. The figure was generated using the program CGView
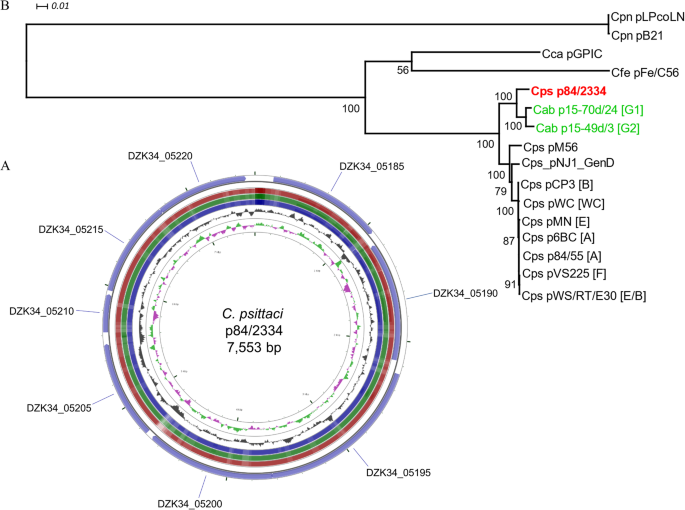
The sequence of the C. psittaci strain 84/2334 plasmid was extracted as a single contig from the total whole genome raw sequence reads by comparison with other C. psittaci plasmid sequences (Table 2). The 84/2334 plasmid comprises a single circular sequence of 7553 bp (Fig. 3a) with eight coding sequences, similar to those found in other sequenced plasmid-bearing chlamydial species [39, 40]. Phylogenetic analysis of the 84/2334 plasmid with other sequenced chlamydial plasmids (including C. psittaci, C. penumoniae, C. felis and C. caviae) shows that it clusters with those of the two avian C. abortus strains (Fig. 3b).
Circular representation of the C. psittaci 84/2334 plasmid sequence and comparative phylogenetic analysis. a Circular representation of the plasmid sequence of strain 84/2334; Circles from the outside in show: the positions of the eight coding sequences on the forward (circle 1) and reverse (circle 2) strands of p84/2334. Circles 3–5 show positions of BLAST hits determined through blastn comparisons of C. psittaci 6BC (circle 3), VS225 (circle 4) and M56 (circle 5) with the following settings: query split size = 50,000 bp, query split overlap size = 0, expect value cut off = 0.0001. The height of the shading in the BLAST results rings is proportional to the percent identity of the hit. Overlapping hits appear as darker shading. Circles 6 and 7 show plots of the GC content and GC skew plotted as the deviation from the average for the entire sequence. The figure was generated using the program CGView. b Phylogenetic tree of an alignment of the plasmid sequences of C. psittaci (Cps), avian C. abortus (Cab), C. caviae (Cca), C. felis (Cfe) and C. pneumoniae (Cpn) strains. The consensus tree was estimated in TOPALi by Maximum Likelihood (PhyML) using a TIM + G substitution and rate heterogeneity model, according to BIC model selection, and 100 non-parametric bootstrap replicates. The tree is midpoint rooted and bootstrap support is indicated by the number at the node. The scale bar indicates the expected substitutions per site. Genotypes are given in square brackets. The tree was prepared in Dendroscope. Strain 84/2334 plasmid is in bold and red font. Avian C. abortus strain plasmids are in green font
Whole genome recombination and phylogenetic analysis
Whole-genome phylogenetic analysis informed by recombination for the genome (chromosomal sequence) of strain 84/2334 compared to other representative C. psittaci and C. abortus strains (Table 2) shows a clear separation of the genomes into two clades, with 84/2334 clustering with the C. abortus clade (Fig. 4). The two avian C. abortus strains 15-59d/3 and 15-70d/24 branch off first from the last common ancestor, with strain 84/2334 branching off second, followed by C. abortus variant strain LLG and then all the remaining C. abortus strains. Additional analysis, adding an outgroup comprising the genome of C. pecorum W73 to the analysis as a control (Additional file 5), conducting the analysis without taking recombination into account (Additional file 6) and NeighborNet analysis (Additional file 7) did not change the structure of the tree and the placement of strain 84/2334 with the C. abortus strains. In contrast to the C. abortus clade, recombination has a greater effect on the C. psittaci clade. For example, while recombination-based analysis identifies strains M56 and NJ1 as the ones branching off first from the C. psittaci last common ancestor (Fig. 4), the two strains populate an internal branch of the tree obtained without taking recombination into account (Additional file 6). It should also be noted that whole-genome phylogenetic analysis without taking recombination into account (Additional file 6) and NeighborNet analysis (Additional file 7) clearly place C. buteonis strain RSHA between the C. psittaci and C. abortus clades.
Whole genome phylogenetic analysis informed by recombination. Phylogenetic tree of a whole genome sequence MAFFT alignment of the C. abortus (Cab) and C. psittaci (Cps) strains shown in Table 2 as derived by Gubbins after removing genomic regions affected by recombination. Genotypes are given in square brackets. FastTree was used as tree builder, and a maximum of 100 iterations was specified in order to guarantee convergence. The tree was midpoint rooted and prepared for publication in Dendroscope. Strain 84/2334 is in bold and red font. Classical and avian C. abortus strains are in blue and green fonts, respectively
Further details on the regions of recombination identified by Gubbins [41] for all of the strains in Table 1 except the two avian C. abortus strains is shown in Fig. 5. The analysis reveals a small number of recombination regions shared within the C. abortus clade and a much larger set common to all the C. psittaci strains. Strain 84/2334 has an intermediate structure, sharing both the regions shared by C. abortus strains and a subset of those common to C. psittaci. Notably, several strains (C. psittaci M56, VS225, and NJ1; and, to a lesser extent, C. psittaci WC and CP3) show greater recombination events unique to the strains. Inclusion of the two avian C. abortus strains in the analysis (Additional file 8) reveals additional regions of recombination that are shared by sequences populating the underlying levels of the tree, with 84/2334 exhibiting a pattern almost identical to that of the non-avian C. abortus strains.
Summary of Gubbins recombination analysis excluding avian C. abortus strains 15-59d/3 and 15-70d/24. The red blocks represent recombination events occurring on an internal branch of the phylogenetic tree, which are shared by several strains by common descent. The blue blocks indicate recombination events occurring on terminal branches of the phylogenetic tree, which are unique to a specific strain. The parameters used for the run are those described in Fig. 4
Comparative genome analysis and large-scale gene variation in strain 84/2334
Comparative analysis of the genome of strain 84/2334 with each of the genomes of the C. abortus and C. psittaci strains in Table 2 shows a high level of sequence conservation, specifically in terms of gene content and order (Fig. 2 and Additional file 9). The level of sequence similarity of strain 84/2334 to the other genomes as indicated by blastn matches visualised using CGView (Fig. 2) and the Artemis Comparison Tool (ACT) (Additional file 9) appears greater with C. psittaci strains 84/55, 6BC, VS225, CP3, NJ1, MN and WC. The main differences occur in the plasticity zone (PZ), polymorphic membrane protein (Pmp) loci and transmembrane-head protein family (TMH) loci of C. psittaci strains GR9, M56 and WS/RT/E30.
Plasticity zone
The PZ of strain 84/2334 most closely resembles that of C. psittaci genotypes A, B, D, E, F and WC in terms of gene content, with the one exception, which is that it does not possess the gene encoding the membrane attack complex/perforin domain-containing protein (MACP) (Fig. 6). MACP is also absent in the PZ region of C. abortus, avian C. abortus and C. psittaci genotypes C, E/B and M56. None of the strains, including 84/2334, encode any of the phospholipase D (PLD) protein family genes or any of the genes involved in L-tryptophan biosynthesis found within or external to the PZ region of some other chlamydial species. Strain 84/2334 is clearly different from the classical C. abortus strains, having an additional six genes, including a large cytotoxin gene (DZK34_02910) and guaB (DZK34_02935) in addition to 4 predicted hypothetical protein genes. However, the large cytotoxin gene is present in the two avian C. abortus strains. Phylogenetic analysis of the predicted protein product of the 84/2334 large cytotoxin gene shows that it clusters with the equivalent predicted proteins of the strains belonging to C. psittaci genotypes A (6BC and 85/55), B (CP3), D (NJ1), E (MN), F (VS225) and WC, while those of the avian C. abortus strains cluster with C. psittaci strains C (GR9), E/B (WS/RT/E30) and M56 (Additional file 10). Indeed, the PZ regions of the two avian C. abortus strains most closely resemble those of these same C. psittaci genotypes (C, E/B, M56), apart from there being fewer of the hypothetical protein genes present between accC and the cytotoxin gene.
Comparative analysis of the genes present in the plasticity zone of C. psittaci strain 84/2334. Comparison of nucleotide matches (computed using blastn) between the genes accB (orange) and guaB (pink) in C. abortus and C. psittaci species and genotypes. Chlamydia psittaci genotypes B, E, F (not shown) are identical in gene content to genotypes A, D and WC. The presence of accC (dark blue), guaA (green), MACP (purple), cytotoxin (red) and hypothetical protein (brown) genes are as indicated. Vertical lines through the arrows indicate point or frame-shift mutations. The orientation of coding sequences in the forward and reverse frames are indicated by the direction of the block arrows. The level of BLAST identity between the sequences is indicated by the degree of grey shading in the vertical bars. The figure was generated using EasyFig
TMH/Inc. protein family
A comparison of the TMH loci of C. abortus and C. psittaci strains shows them to have a high degree of similarity, with the majority of the C. psittaci strains (from genotypes A, B, E, F, WC) carrying all 11 of the genes found in C. abortus, differing only in pseudogene content (Fig. 7). Strain 84/2334 is also very similar in content to these strains/genotypes other than lacking the CAB766 gene (from C. abortus strain S26/3; ortholog CPSIT_0846 in C. psittaci 6BC). In contrast, significant differences were noted for C. psittaci strains GR9 and WS/RT/E30, which have a smaller TMH locus with considerably fewer intact genes plus many gene remnants (Fig. 7). The two avian C. abortus strains (15-49d/3 and 15-70d/24) do not show homology in this region with the typical C. abortus strains or indeed strain 84/2334, rather they are most similar to the smaller TMH loci of C. psittaci strains GR9 and WS/RT/E30.
Comparative analysis of the genes present in the transmembrane head of C. psittaci strain 84/2334. Comparison of nucleotide matches (computed using blastn) between the genes lpxB and pmpD (both orange) in C. abortus and C. psittaci species and genotypes. Chlamydia psittaci genotypes B, E, F (not shown) are identical in gene content to genotypes A, D and WC. The presence of TMH/Inc. (green) and hypothetical protein genes/gene remnants (brown) are as indicated. The 3 genes coding for proteins of unknown function are also indicated (blue). Vertical lines through the arrows indicate point or frame-shift mutations. The orientation of coding sequences in the forward and reverse frames are indicated by the direction of the block arrows. The level of BLAST identity between the sequences is indicated by the degree of grey shading in the vertical bars. The figure was generated using EasyFig
Polymorphic membrane protein family
A total of 20 Pmps were identified in strain 84/2334 through comparative comparison with the other strains and through searching for classical Pmp motifs and sequence secondary structure predictions, specifically for a N-terminal domain containing repeat GG[A/L/V/I][I/L/V/Y] and FXXN, a central PmpM domain, a C-terminal autotransporter (AT) domain and a terminal phenylalanine amino acid [42, 43] (Fig. 8). The genes were located in four loci, with single members of the PmpA, PmpB, PmpD and PmpH families and expansion of the PmpE and PmpG families identified, as observed in the other species/strains [42,43,44,45,46,47]. Following artificial reconstruction of predicted pseudogenes, phylogenetic analysis of the identified sequences confirmed our assignment of the identified Pmps to specific families (Additional file 11). This analysis also showed that strain 84/2334 Pmps from each of the A, B, D, E and H families clustered with the C. abortus strains, with the exception of PmpE2 which clustered more closely with the C. psittaci clade along with the equivalent proteins of the two avian C. abortus strains. Considerable expansion in the PmpG family was observed for strain 84/2334, with a total of 13 Pmps identified (five of which are pseudogenes) (Fig. 8). The two additional Pmps observed in strain 84/2334 (G7 and G12 in Fig. 8) compared to the classical C. abortus strains were determined to be gene duplications of two of the PmpG family members that carry homopolymeric (nucleotide ‘G’) tracts. One of these duplications (PmpG12) present in locus 3 (Fig. 8) is a pseudogene, where the other two PmpG proteins (PmpG11 and PmpG13), from which comparative genome analysis suggests it has arisen, are also pseudogenes. The frame-shifts in all three genes occur in homopolymeric (nucleotide ‘G’) tracts. The other gene duplication (PmpG7) in locus 2, as well as PmpG6 (intact gene) and PmpG8 (pseudogene) from which comparative analysis suggests the duplication has arisen, all similarly possess homopolymeric ‘G’ tracts.
Polymorphic membrane proteins in C. psittaci strain 84/2334. a Gene families and gene organisation in the four Pmp loci (as indicated) showing intact genes (orange) and pseudogenes (grey), identified following BLAST and phylogenetic comparison with other published Pmps. Pmp gene designations are indicated under each block arrow. b Schematic diagram showing the conserved Pmp features, comprising the predicted PmpM domains (blue rectangular block), autotransporter domains (red rectangular block) and pmp passenger domain repeat motifs GG[A/L/V/I][I/L/V/Y] (blue vertical lines) and FXXN (red vertical lines) (Only motifs in which FXXN follows GG[A/L/V/I][I/L/V/Y] are shown). The predicted number of amino acids (aa) is indicated to the right of each gene. Pmp gene identification numbers and gene identification tags (indicated as DZK34_0xxxx) are indicated to the left of each gene
Discussion
In this study, application of a previously developed MLST typing system [32], as well as cluster analysis of MLST allelic profiles/STs, extends previous observations showing clustering of the 84/2334 strain with classical C. abortus strains [28] by also showing clustering with the avian C. abortus strains (15-70d/24, 15-49d/3 and 15-58d/44). These results were further supported by rMLST, in silico genome-to-genome distance comparisons and 16S rRNA gene phylogenetic analysis, as well as by analysis utilising the recently developed Pillonel typing system [35], all showing similar clustering of the strain with C. abortus species, and closer to classical than avian strains. Interestingly, although not a focus of this study, MLST, rRNA, whole genome and NeighborNet phylogenetic analyses all clearly show that C. buteonis is phylogenetically positioned between both C. psittaci and C. abortus species, as previously suggested [7], and although shares similarities with both, appears to cluster more closely with C. psittaci and particularly strain M56.
To characterise strain 84/2334 further we sequenced the whole genome, including plasmid sequence, for a more detailed comparison with previously sequenced representative strains of C. abortus and C. psittaci genotypes [16, 27, 42, 48,49,50]. While the chromosomal genome characteristics were essentially similar to those of both C. abortus and C. psittaci species, the GC content was identical to that of C. abortus rather than C. psittaci species, again suggesting greater similarity with C. abortus. However, strain 84/2334 carries an extrachromosomal plasmid, which is more in keeping with C. psittaci species. Chlamydial plasmids, which are non-conjugative and non-integrative (with the exception of tetracycline resistance in C. suis), have been found in most chlamydial species, including C. psittaci, but to date none have been found in classical C. abortus species [51, 52]. However, there have been recent reports of strains carrying plasmids that have been classified as avian C. abortus strains [18, 49, 50]. Interestingly, the 84/2334 plasmid sequence appears more closely related to these avian C. abortus strains than to those of the C. psittaci genotypes, perhaps reflecting an ancestral relationship with classical C. abortus strains that have lost their plasmid through a process of reductive evolution, where in their specific niche, in a principally mammalian livestock host, the plasmid is not required for tropism and pathogenesis.
The designation of strain 84/2334 is further supported by whole genome phylogenetic analysis informed by recombination using Gubbins [41], which uses SNP density to identify recombination blocks. This analysis places strain 84/2334 firmly within the C. abortus clade, in agreement with the typing analyses and also classifies it as an intermediate between the avian and variant/classical C. abortus strains, with branch lengths suggesting a closer evolutionary relationship with classical C. abortus strains. Overall, strain 84/2334 appears much closer to the C. abortus last common ancestor than to the two avian C. abortus strains. Interestingly, recombination has the greatest effect on the phylodynamics of the C. psittaci clade, whereby the placement of strains within the tree structure significantly differs depending on whether phylogenetics is informed or not by recombination. These findings are consistent with previous studies showing that the level of diversity within classical C. abortus strains is low, with only 724 SNPs within the major C. abortus clade and 6718 variable sites within the whole phylogeny (including the variant strain LLG/POS clade) [27], while diversity is much greater in C. psittaci with 47,710 variable sites [19]. Overall, the greater accuracy of the phylogenetic tree obtained when taking recombination into account was confirmed by whole genome NeighborNet network analysis, which produced results essentially identical to those of Gubbins for both the 84/2334 and the C. psittaci strains.
Thus, all the analyses, both with and without taking recombination into account, agree with 84/2334 being a misclassified C. abortus strain that branched off from the C. abortus last common ancestor at some point between the avian and the classical C. abortus strains. This was supported further through a more detailed analysis of the regions of recombination identified by Gubbins, with strain 84/2334 having an intermediate structure, comprising a couple of regions shared by C. abortus strains plus a subset of those common to C. psittaci, while inclusion of the avian C. abortus strains 15-59d/3 and 15-70d/24 added additional regions of recombination, resulting in a distinctively different pattern and again pointing to 84/2334 being closer in evolution to the non-avian C. abortus strains.
The genome of strain 84/2334 shares characteristic synteny in terms of gene content and order with the classical C. abortus, avian C. abortus and C. psittaci strains included in this study, as has been observed for other sequenced chlamydial species [42, 43, 53, 54]. However, differences were observed in a number of genes/loci found in regions of extensive variation that have been suggested to be associated with virulence, niche specificity and disease pathogenesis, particularly the PZ, TMH and Pmp loci.
The PZ region, which is defined as bounded by genes inosine-5′-monophosphate dehydrogenase (guaB) and acetyl-CoA carboxylase (accB), has been found to vary markedly in sequence size (approx. 12–82 kb) and gene content (11–44) across chlamydial species [10, 43]. This highly variable region has been found to carry genes encoding proteins involved in carbohydrate/lipid metabolism (accBC), purine metabolism (guaAB-add) and tryptophan biosynthesis (trpABFCDR, kynU, prsA), as well as genes encoding a membrane attack complex/perforin domain-containing protein (MACP), a phospholipase D (PLD) family of proteins, a varying number of cytotoxin/adherence factor proteins and a number of hypothetical proteins of unknown function. Chlamydia abortus has one of the smallest PZ regions amongst chlamydial species, spanning approx. 12 kb, while the PZ of C. psittaci spans approx. 21–30 kb depending on genotype. While these regions differ extensively between C. abortus and C. psittaci, no variation in gene order or content within C. abortus species has been observed [27]. In contrast, within C. psittaci species there are large differences in gene content at these locations in the different genotypes, perhaps reflecting wide-ranging differences in host specificity. The PZ of strain 84/2334, in keeping with classical C. abortus, avian C. abortus and C. psittaci strains does not contain any of the genes involved in L-tryptophan biosynthesis found within or external to the PZ region of some other chlamydial genomes. Overall, while the PZ of 84/2334 is most similar to that of C. psittaci possessing an additional number of similar hypothetical proteins and a cytotoxin gene, it lacks the MACP gene, which is also absent in C. abortus. MACP has been suggested to be involved in assisting PLDs in lipid acquisition and processing [55], while no PLD proteins were similarly identified in 84/2334 or indeed any of the strains. Although the function of the cytotoxin is unclear, related cytotoxins in E. coli and C. difficile have a role in glycosylation of Rho and Ras GTP-binding proteins, inhibiting lymphocyte activation, host signalling and blocking the induction of interferon-gamma (IFN-γ) [56, 57]. As IFN-γ decreases the availability of L-tryptophan and can lead to resolution of chlamydial infections [58, 59], the ability to block its production may be an important virulence determinant that allows the organism to form persistent subclinical infections, particularly when these pathogens appear to lack the ability to synthesise tryptophan with the absence of the tryptophan biosynthesis operon. However, it is unclear whether the encoded cytotoxin in strain 84/2334 is actively expressed or truncated at the amino terminus as a number of frameshift mutations are present at the 5′ end of the gene. But this gene is present in all of the C. psittaci strains/genotypes, including the avian C. abortus strains, although phylogenetic analysis shows that it clusters with different C. psittaci genotypes than the avian C. abortus strains, perhaps reflecting a different evolutionary path and the ultimate loss of this gene in classical C. abortus strains.
Another region that has been identified as important in terms of virulence is the TMH locus that typically has 11 genes between the lipid A disaccharide synthase gene (lpxB) and the polymorphic membrane protein D (pmpD) [42] that encode proteins with either paired (n = 8) or single (n = 3) N-terminal transmembrane domains followed by alpha-helical coiled-coil domains of varying lengths, with an amino acid composition rich in leucine, glutamate and serine residues or contain conserved domains of unknown function [42, 48]. These proteins, which lack a signal sequence, are thought to be secreted via the Type III Secretion System and have been associated with the chlamydial inclusion membrane, chlamydial growth and host inflammatory responses [60,61,62]. They are also thought to be related to the Inc-protein family, which similarly possess no primary sequence similarity but have unique paired hydrophobic domains in either the N-terminal (IncA) or C-terminal (IncB and IncC) regions (all three Inc. proteins are present in strain 84/2334 and all the other C. abortus and C. psittaci strains included in this study) [27, 44, 48]. Although there are few differences in gene content between strain 84/2334 and C. abortus and most C. psittaci genotypes, with some differences in pseudogene content, the main difference is in the absence of the CAB766 gene in 84/2334. This gene encodes the only TMH protein, termed Inc766, to be characterised from this region to date, and which has been shown to form oligomers and to be localised to the extra-inclusion space [63]. This possibly suggests a role for this protein and other TMH proteins in vesicular trafficking and modulation of host cell functions, as suggested for other Incs [60, 63, 64]. In contrast, the TMH region for the two avian C. abortus strains and C. psittaci strains GR9 and WS/RT/E30 is much smaller with considerably fewer intact genes plus many gene remnants, again perhaps reflecting a different evolutionary process for these avian strains compared to strain 84/2334.
A major source of large-scale variation among Chlamydia spp. is in the Pmp proteins, which are members of the type V “autotransporter (AT)” secretion system [65]. Although the function of these proteins has largely to be determined they have been suggested to be involved in niche adaptation and host immune evasion, with specific functions on host cell adherence, molecular transport and cell wall associated functions [66,67,68]. The number of Pmps in the family Chlamydiaceae has been shown to vary considerably from 9 to 21 depending on species, with C. abortus having 18 and C. psittaci 17–21 [42, 44,45,46,47, 53, 69]. These Pmps are broadly classified into six families (A, B/C, D, E/F, G/I, and H) that share little primary sequence homology but which have distinct characteristic features, possessing an N-terminal passenger domain with a variable number of repeat motifs GG[A/L/V/I][I/L/V/Y] and FXXN, a central PmpM domain, a C-terminal AT domain and all end with the amino acid phenylalanine. In contrast, Pmps within a family share primary sequence homology both within and across species/strains. In this study we identified a total of 20 Pmps in strain 84/2334, with phylogenetic analysis showing that individual family members cluster more closely with the equivalent proteins from C. abortus, suggesting a closer evolutionary relationship. The additional two Pmps in 84/2334 to those found in C. abortus strains result from gene duplication and expansion of the G family from 11 to 13, similar to the number observed in C. psittaci (n = 10–14). However, these gene duplications and the genes they have arisen from possess homopolymeric tracts that are thought to be subject to phase variation by slip-strand mispairing [42]. These observations, coupled with the expansion and variation observed in this PmpG family across the C. abortus and C. psittaci species/strains, highlights that this is the most diverse and rapidly evolving of all the Pmp families, likely arising and evolving as a result of gene duplications and losses in the various chlamydial ancestries and indicative of the observed recombinogenic nature of this region of the genome [27, 42].
Conclusion
In this study we have fully characterised the C. psittaci strain 84/2334 at the whole genome level using existing typing systems, as well as through whole genome sequencing, comparative genomics and recombination analysis. These analyses show an evolutionary relationship between the strain with both C. abortus and C. psittaci species, in agreement with a previous publication suggesting the strain to be a missing link between C. psittaci and C. abortus and classifying it as an intermediary strain [21]. However, this study shows that the strain is not intermediary at all, but rather much closer in evolution to the classical non-avian C. abortus strains than to the avian C. abortus strains or C. psittaci species, branching off from the last common ancestor of C. abortus between the avian and classical strains. These results coupled with the presence of the plasmid sequence which clusters more closely with the avian C. abortus plasmids than the C. psittaci or other chlamydial plasmids, along with taking into account similarities and differences in gene content in key regions and loci associated with virulence, niche specificity and pathogenesis are all consistent with strain 84/2334 being an ancestral C. abortus species. Therefore, overall, the results of this study support the reclassification of this strain as C. abortus species. Future studies should aim to characterise the strain further, investigating its pathogenesis in challenge model systems, as well as with the recent advances in plasmid-based transformation of chlamydial species [70, 71] begin to understand gene function as well as look towards the future development of new novel vaccines.
Methods
Cell culture, DNA extraction and genome sequencing
Chlamydia psittaci strain 84/2334, isolated from the lungs of an imported yellow-headed Amazon parrot in Germany [31], was grown in Buffalo Green Monkey kidney (BGM; obtained from ECACC General Collection, product number 90092601) cells using standard techniques [72]. Infected cells were harvested to purify chlamydial elementary bodies, as previously published [73], and DNA extracted using a DNeasy Blood & Tissue kit (Qiagen Ltd., Belgium). Genomic DNA was fractionated into smaller fragments (300–800 bp), blunt-ended, adapters ligated onto fragments and attached to DNA-capture beads to generate a single-stranded template library for pyrosequencing on a 454 GS-FLX Titanium (Roche) pyrosequencer, according to the manufacturer’s instructions.
Mapping, assembly, annotation and comparative analysis
Following quality filtering, de novo assembly and reference mapping assembly to each of the C. psittaci genomes in Table 1 was performed using the software program Newbler (version 2.3; 454 Life Sciences, Branford, CT, USA). De novo assembly produced a single contig, with 273x sequencing coverage. Genome annotation was performed using the NCBI prokaryotic genome annotation pipeline (PGAP) [74] and manually curated using Artemis [75]. Pseudogenes were defined as having one or more mutations that would ablate expression (i.e. indel or substitution causing frameshift or stop codon). The origin of replication was determined using Ori-Finder [76] and the genome adjusted so that the first base was upstream of hemB in the oriC region. To identify any reads mapping to a plasmid, the plasmids of C. psittaci strains 6BC and 84/55 (Table 2) were used as reference sequences.
Comparative genome and plasmid analyses of 84/2334 was performed against representative strains of each C. psittaci genotype (6BC, 84/55, CP3, GR9, NJ1, MN, VS225, M56, WC [16]), C. abortus species (S26/3 [42], AB7 [77], LLG [27, 48]) and avian C. abortus strains (genotypes G1 and G2 [18, 50]). For the plasmid comparisons, sequences were oriented to start at a position relative to nucleotide 6848 in strain 84/2334. Global genome and plasmid comparisons were visualised using the Artemis Comparison Tool (ACT) [78] with crunch input files generated by running pairwise blastn comparisons of the sequences using Megablast within the ncbi-blast-2.9.0+ command line software [79]. Genome and plasmid maps were generated using the CGView server [80].
Molecular genotyping schemes
MLST genotyping of gene fragments of seven housekeeping genes (enoA, fumC, gatA, gidA, hemN, hlfX and oppA) of C. psittaci strain 84/2334 and representative strains from Chlamydiaceae species, was performed as previously described [28, 32]. Genes were concatenated, aligned using MAFFT and a phylogenetic tree estimated in TOPALi as detailed in the following section (Phylogenetic and network analysis). Cluster analysis based on defined allelic profiles or sequence types (ST) for each individual isolate, as well as STs from other representative isolates for all Chlamydiaceae species (except C. trachomatis) in the online database [36, 81] was conducted to generate minimum spanning trees that were visualised using GrapeTree [82]. Ribosomal MLST analysis to determine species classification was conducted using the rps gene database [34, 83]. Digital DNA-DNA hybridization to determine species was conducted using the Genome-to-Genome Distance Calculator 2.1 [33, 84].
A classification scheme, based on five discriminant proteins (Adk, FtsK, HemL, PepF and RpoN) for species designation, was applied to strain 84/2334, as previously described [35]. Protein sequences for all strains were identified by BLAST analysis. Sequence distances (% ID) for each of the five proteins from strain 84/2334 with each of the equivalent proteins from the C. psittaci and C. abortus strains and genotypes in Table 2 were calculated by aligning sequences in MegAlign 15 (Lasergene software, DNASTAR Inc., Madison, WI, USA) Clustal Omega [85]. Proteins with a %ID ≥95% for Adk and HemL, ≥ 96% for PepF and RpoN, and ≥ 98% for FtsK, indicate they are classified as the same species.
Phylogenetic and network analysis
Whole genome, plasmid, gene (16S rRNA, concatenated MLST housekeeping genes) and protein (Pmps, cytotoxins) sequences were aligned using MAFFT v7.450 [86] or Clustal Omega. Phylogenetic trees were estimated in IQ-Tree v2.0.5 [87] or TOPALi v2.5 [88] by Maximum Likelihood using GTR + G + I (concatenated MLST housekeeping genes), TVM + F + R6 (genomes), TIM + G (plasmids), JTTDCMut + F + I + G4 (Cytotoxins; PmpsABDH), JTT + F + R3 (PmpsE) and JTT + F + R5 (PmpsG) substitution and rate models or by NeighbourJoining using F84 + G (16S rRNA genes) substitution and rate models. Models were selected according to Bayesian information criterion (BIC). Bootstrap analyses were performed on 1000 replicate trees. All trees were midpoint rooted, unless otherwise indicated in the figure legend. Trees were prepared for publication using Dendroscope 3 [89]. Phylogenetic network analysis for inferring evolutionary relationships between the MAFFT aligned genome species and strains was performed using SplitsTree v4.15.1 [90].
Recombination analysis
To investigate the effect of recombination on the phylogeny Gubbins version 2.4.1 [41] was used. Genome sequences (Table 2) were aligned using MAFFT v7.471 [86]. Recombination analysis was performed with and without the two avian C. abortus strains (15-70d/24 and 15-49d/3) present in the alignments.
Plasticity zone, transmembrane head and polymorphic membrane protein gene analysis
The PZ region, spanning genes between guaB and accB, and TMH/Inc. locus, typically encoding genes between lpxB and pmpD were manually identified in each of the C. abortus and C. psittaci genomes using ACT [78]. BLAST searches were also performed to confirm gene content within these loci. PZ and TMH/Inc. loci were extracted for each of the genomes and linear comparisons were produced using Easyfig for Windows version 2.2.5 [91].
Gene sequences for the polymorphic membrane proteins (Pmps) were manually checked in each of the C. abortus and C. psittaci genomes using ACT [78] to determine functionality and identify pseudogenes i.e. where there were one or more mutations that would ablate expression. For the purposes of this study all identified pseudogenes were artificially constructed in silico by comparison with intact homologous genes in other strains/species so that they could be included in the analysis. Prior to the phylogenetic analysis of Pmp gene families detailed in the previous section (Phylogenetic and network analysis), the Pmp protein sequences were initially classified into their respective family groups (A, B, D, E, G and H) through phylogenetic analysis of a single alignment of all the sequences in IQ-Tree. Pmp-specific C-terminal autotransporter β-barrel domains and the conserved PmpM middle domain motifs were identified using the Pfam HMM database [92]. Predicted Pmp passenger domain repeat motifs were identified manually where GG[A/L/V/I][I/L/V/Y] motifs were closely followed by an FXXN motif.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its additional files) and in the NCBI repository under BioProject accession PRJNA485898. The complete assemblies of both the chromosomal and plasmid sequences are available under accession numbers CP031646 and CP031647, respectively.
The dataset(s) supporting the conclusions of this article is (are) available in the [repository name] repository, [unique persistent identifier and hyperlink to dataset(s) in http:// format].
Abbreviations
- ACT:
-
Artemis comparison tool
- AT:
-
Autotransporter
- EAE:
-
Enzootic abortion of ewes
- IFN-γ:
-
Interferon-gamma
- MACP:
-
Membrane attack complex protein
- MLST:
-
Multi-locus sequence typing
- OEA:
-
Ovine enzootic abortion
- PLD:
-
Phospholipase D
- Pmp:
-
Polymorphic membrane protein
- PZ:
-
Plasticity zone
- rMLST:
-
Ribosomal multi-locus sequence typing
- SNP:
-
Single nucleotide polymorphism
- TMH:
-
Transmembrane head
References
Longbottom D, Coulter LJ. Animal chlamydioses and zoonotic implications. J Comp Pathol. 2003;128:217–44.
Everett KDE, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of ParaChlamydiaceae fam. Nov. and Simkaniaceae fam. Nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49(Pt2):415–40.
Stephens RS, Myers G, Eppinger M, Bavoil PM. Divergence without difference: phylogenetics and taxonomy of Chlamydia resolved. FEMS Immunol Med Microbiol. 2009;55(2):115–9.
Schachter J, Stephens RS, Timms P, Kuo C, Bavoil PM, Birkelund S, et al. Radical changes to chlamydial taxonomy are not necessary just yet. Int J Syst Evol Microbiol. 2001;51(Pt 1):249.
Horn M. Phylum XXIV. Chlamydiae Garrity and Holt 2001. In: Krieg NR, Staley JT, Brown DR, Hedlund BP, Paster BJ, Ward NL, Ludwig W, Whitman WB, editors. Bergey's manual of systematic bacteriology. 4. 2nd ed. New York: Springer; 2011. p. 843–78.
Sachse K, Bavoil PM, Kaltenboeck B, Stephens RS, Kuo CC, Rossello-Mora R, et al. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst Appl Microbiol. 2015;38(2):99–103.
Laroucau K, Vorimore F, Aaziz R, Solmonson L, Hsia RC, Bavoil PM, et al. Chlamydia buteonis, a new Chlamydia species isolated from a red-shouldered hawk. Syst Appl Microbiol. 2019;42(5):125997.
Staub E, Marti H, Biondi R, Levi A, Donati M, Leonard CA, et al. Novel Chlamydia species isolated from snakes are temperature-sensitive and exhibit decreased susceptibility to azithromycin. Sci Rep. 2018;8(1):5660.
Vorimore F, Hsia RC, Huot-Creasy H, Bastian S, Deruyter L, Passet A, et al. Isolation of a new Chlamydia species from the feral sacred Ibis (Threskiornis aethiopicus): Chlamydia ibidis. PLoS One. 2013;8(9):e74823.
Taylor-Brown A, Bachmann NL, Borel N, Polkinghorne A. Culture-independent genomic characterisation of Candidatus chlamydia sanzinia, a novel uncultivated bacterium infecting snakes. BMC Genomics. 2016;17:710.
Taylor-Brown A, Spang L, Borel N, Polkinghorne A. Culture-independent metagenomics supports discovery of uncultivable bacteria within the genus Chlamydia. Sci Rep. 2017;7(1):10661.
Laroucau K, Ortega N, Vorimore F, Aaziz R, Mitura A, Szymanska-Czerwinska M, et al. Detection of a novel Chlamydia species in captive spur-thighed tortoises (Testudo graeca) in southeastern Spain and proposal of Candidatus chlamydia testudinis. Syst Appl Microbiol. 2020;43(2):126071.
Harkinezhad T, Geens T, Vanrompay D. Chlamydophila psittaci infections in birds: a review with emphasis on zoonotic consequences. Vet Microbiol. 2009;135(1–2):68–77.
Sillis M, Longbottom D. Chlamydiosis. In: Palmer SR, Soulsby L, Torgerson PR, Brown DWG, editors. Zoonoses. 2nd ed. Oxford: Oxford University Press; 2011. p. 146–57.
Geens T, Desplanques A, Van Loock M, Bonner BM, Kaleta EF, Magnino S, et al. Sequencing of the Chlamydophila psittaci ompA gene reveals a new genotype, E/B, and the need for a rapid discriminatory genotyping method. J Clin Microbiol. 2005;43(5):2456–61.
Van Lent S, Piet JR, Beeckman D, van der Ende A, Van Nieuwerburgh F, Bavoil P, et al. Full genome sequences of all nine Chlamydia psittaci genotype reference strains. J Bacteriol. 2012;194(24):6930–1.
Sachse K, Laroucau K, Hotzel H, Schubert E, Ehricht R, Slickers P. Genotyping of Chlamydophila psittaci using a new DNA microarray assay based on sequence analysis of ompA genes. BMC Microbiol. 2008;8:63.
Szymanska-Czerwinska M, Mitura A, Niemczuk K, Zareba K, Jodelko A, Pluta A, et al. Dissemination and genetic diversity of chlamydial agents in polish wildfowl: isolation and molecular characterisation of avian Chlamydia abortus strains. PLoS One. 2017;12(3):e0174599.
Read TD, Joseph SJ, Didelot X, Liang B, Patel L, Dean D. Comparative analysis of Chlamydia psittaci genomes reveals the recent emergence of a pathogenic lineage with a broad host range. MBio. 2013;4(2):e00604–12.
Sachse K, Laroucau K, Riege K, Wehner S, Dilcher M, Creasy HH, et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp nov and Chlamydia gallinacea sp nov. Syst Appl Microbiol. 2014;37(2):79–88.
Van Loock M, Vanrompay D, Herrmann B, Vander Stappen J, Volckaert G, Goddeeris BM, et al. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int J Syst Evol Microbiol. 2003;53(Pt 3):761–70.
Aitken ID, Longbottom D. Chlamydial abortion. In: Aitken ID, editor. Diseases of sheep. Fourth ed. Oxford: Blackwell Publishing; 2007. p. 105–11.
Stamp JT, McEwen AD, Watt JAA, Nisbet DI. Enzootic abortion in ewes. I. Transmission of the disease. Vet Rec. 1950;62:251–4.
Buxton D. Potential danger to pregnant women of Chlamydia psittaci from sheep. Vet Rec. 1986;118:510–1.
Pospischil A, Thoma R, Hilbe M, Gresi P, Gebbers JO. Abortion in woman caused by caprine Chlamydophila abortus (Chlamydia psittaci serovar 1). Swiss Med Wkly. 2002;132(5–6):64–6.
Essig A, Longbottom D. Chlamydia abortus: new aspects of infectious abortion in sheep and potential risk for pregnant women. Curr Clin Microbiol. 2015;2(1):22–34.
Seth-Smith HMB, Buso LS, Livingstone M, Sait M, Harris SR, Aitchison KD, et al. European Chlamydia abortus livestock isolate genomes reveal unusual stability and limited diversity, reflected in geographical signatures. BMC Genomics. 2017;18(1):344.
Pannekoek Y, Dickx V, Beeckman DS, Jolley KA, Keijzers WC, Vretou E, et al. Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS One. 2010;5(12):e14179.
Fukushi H, Nojiri K, Hirai K. Monoclonal antibody typing of Chlamydia psittaci strains derived from avian and mammalian species. J Clin Microbiol. 1987;25(10):1978–81.
Sayada C, Andersen AA, Storey C, Milon A, Eb F, Hashimoto N, et al. Usefulness of omp1 restriction mapping for avian Chlamydia psittaci isolate differentiation. Res Microbiol. 1995;146(2):155–65.
Vanrompay D, Butaye P, Sayada C, Ducatelle R, Haesebrouck F. Characterization of avian Chlamydia psittaci strains using omp1 restriction mapping and serovar-specific monoclonal antibodies. Res Microbiol. 1997;148(4):327–33.
Pannekoek Y, Morelli G, Kusecek B, Morre SA, Ossewaarde JM, Langerak AA, et al. Multi locus sequence typing of Chlamydiales: clonal groupings within the obligate intracellular bacteria Chlamydia trachomatis. BMC Microbiol. 2008;8:42.
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60.
Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM, et al. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology. 2012;158(Pt 4):1005–15.
Pillonel T, Bertelli C, Salamin N, Greub G. Taxogenomics of the order Chlamydiales. Int J Syst Evol Microbiol. 2015;65(Pt 4):1381–93.
Pannekoek Y. PubMLST: Public databases for molecular typing and microbial genome diversity - Chlamydiales spp. [Available from: https://pubmlst.org/organisms/chlamydiales-spp. Accessed 30 November 2020].
Rodolakis A. In vitro and in vivo properties of chemically induced temperature-sensitive mutants of Chlamydia psittaci var. ovis: screening in a murine model. Infect Immun. 1983;42(2):525–30.
Livingstone M, Wheelhouse N, Ensor H, Rocchi M, Maley S, Aitchison K, et al. Pathogenic outcome following experimental infection of sheep with Chlamydia abortus variant strains LLG and POS. PLoS One. 2017;12(5):e0177653.
Thomas NS, Lusher M, Storey CC, Clarke IN. Plasmid diversity in Chlamydia. Microbiology. 1997;143(Pt 6):1847–54.
Seth-Smith HM, Harris SR, Persson K, Marsh P, Barron A, Bignell A, et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics. 2009;10:239.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15.
Thomson NR, Yeats C, Bell K, Holden MT, Bentley SD, Livingstone M, et al. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 2005;15(5):629–40.
Sait M, Livingstone M, Clark EM, Wheelhouse N, Spalding L, Markey B, et al. Genome sequencing and comparative analysis of three Chlamydia pecorum strains associated with different pathogenic outcomes. BMC Genomics. 2014;15:23.
Voigt A, Schofl G, Saluz HP. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS One. 2012;7(4):e35097.
Read TD, Myers GS, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, et al. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 2003;31(8):2134–47.
Mitchell CM, Hovis KM, Bavoil PM, Myers GS, Carrasco JA, Timms P. Comparison of koala LPCoLN and human strains of Chlamydia pneumoniae highlights extended genetic diversity in the species. BMC Genomics. 2010;11:442.
Grimwood J, Stephens RS. Computational analysis of the polymorphic membrane protein superfamily of Chlamydia trachomatis and Chlamydia pneumoniae. Microb Comp Genomics. 1999;4(3):187–201.
Sait M, Clark EM, Wheelhouse N, Livingstone M, Spalding L, Siarkou VI, et al. Genome sequence of the Chlamydophila abortus variant strain LLG. J Bacteriol. 2011;193(16):4276–7.
Szymańska-Czerwińska MZK, Mitura A, Laroucau K, Niemczuk K, editors. Whole-genome sequencing of avian Chlamydia abortus strains isolated from wild birds. Odessa, Ukraine: Fifth European Meeting on Animal Chlamydioses; 2018. p. 35–7.
Zareba-Marchewka K, Szymanska-Czerwinska M, Mitura A, Niemczuk K. Draft genome sequence of avian Chlamydia abortus genotype G1 strain 15-70d24, isolated from Eurasian teal in Poland. Microbiol Resour Announc. 2019;8:e00658–19.
Pawlikowska-Warych M, Sliwa-Dominiak J, Deptula W. Chlamydial plasmids and bacteriophages. Acta Biochim Pol. 2015;62(1):1–6.
Joseph SJ, Marti H, Didelot X, Castillo-Ramirez S, Read TD, Dean D. Chlamydiaceae genomics reveals interspecies admixture and the recent evolution of Chlamydia abortus infecting lower mammalian species and humans. Genome Biol Evol. 2015;7(11):3070–84.
Read TD, Brunham RC, Shen C, Gill SR, Heidelberg JF, White O, et al. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 2000;28(6):1397–406.
Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, et al. Unity in variety--the pan-genome of the Chlamydiae. Mol Biol Evol. 2011;28(12):3253–70.
Nunes A, Gomes JP. Evolution, phylogeny, and molecular epidemiology of Chlamydia. Infect Genet Evol. 2014;23:49–64.
von Eichel-Streiber C, Boquet P, Sauerborn M, Thelestam M. Large clostridial cytotoxins--a family of glycosyltransferases modifying small GTP-binding proteins. Trends Microbiol. 1996;4(10):375–82.
Klapproth JM, Scaletsky IC, McNamara BP, Lai LC, Malstrom C, James SP, et al. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect Immun. 2000;68(4):2148–55.
Beatty WL, Byrne GI, Morrison RP. Repeated and persistent infection with Chlamydia and the development of chronic inflammation and disease. Trends Microbiol. 1994;2(3):94–8.
Taylor MW, Feng GS. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. 1991;5(11):2516–22.
Gitsels A, Sanders N, Vanrompay D. Chlamydial infection from outside to inside. Front Microbiol. 2019;10:2329.
Dehoux P, Flores R, Dauga C, Zhong G, Subtil A. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics. 2011;12:109.
Pannekoek Y, van der Ende A. Inclusion proteins of Chlamydiaceae. Drugs Today (Barc). 2006;42(Suppl A):65–73.
Vretou E, Katsiki E, Psarrou E, Vougas K, Tsangaris GT. Identification and characterization of Inc766, an inclusion membrane protein in Chlamydophila abortus-infected cells. Microb Pathog. 2008;45(4):265–72.
Brumell JH, Scidmore MA. Manipulation of Rab GTPase function by intracellular bacterial pathogens. Microbiol Mol Biol Rev. 2007;71(4):636–52.
Henderson IR, Lam AC. Polymorphic proteins of Chlamydia spp. - autotransporters beyond the Proteobacteria. Trends Microbiol. 2001;9(12):573–8.
Molleken K, Becker E, Hegemann JH. The Chlamydia pneumoniae invasin protein Pmp21 recruits the EGF receptor for host cell entry. PLoS Pathog. 2013;9(4):e1003325.
Vasilevsky S, Stojanov M, Greub G, Baud D. Chlamydial polymorphic membrane proteins: regulation, function and potential vaccine candidates. Virulence. 2016;7(1):11–22.
Gomes JP, Nunes A, Bruno WJ, Borrego MJ, Florindo C, Dean D. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol. 2006;188(1):275–86.
Bachmann NL, Polkinghorne A, Timms P. Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol. 2014;22(8):464–72.
Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR. Clarke IN development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 2011;7(9):e1002258.
Shima K, Weber MM, Schnee C, Sachse K, Käding N, Klinger M, Rupp J. Development of a plasmid shuttle vector system for genetic manipulation of Chlamydia psittaci. mSphere. 2020;5:e00787–20.
Vanrompay D, Ducatelle R, Haesebrouck F. Diagnosis of avian chlamydiosis: specificity of the modified Gimenez staining on smears and comparison of the sensitivity of isolation in eggs and three different cell cultures. Zentralbl Veterinarmed B. 1992;39(2):105–12.
Vanrompay D, Charlier G, Ducatelle R, Haesebrouck F. Ultrastructural changes in avian Chlamydia psittaci serovar A-, B-, and D-infected Buffalo green monkey cells. Infect Immun. 1996;64(4):1265–71.
Galens K, Orvis J, Daugherty S, Creasy HH, Angiuoli S, White O, et al. The IGS standard operating procedure for automated prokaryotic annotation. Stand Genomic Sci. 2011;4(2):244–51.
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16(10):944–5.
Gao F, Zhang CT. Ori-finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics. 2008;9:79.
Longbottom D, Sait M, Livingstone M, Laroucau K, Sachse K, Harris SR, et al. Genomic evidence that the live Chlamydia abortus vaccine strain 1B is not attenuated and has the potential to cause disease. Vaccine. 2018;36(25):3593–8.
Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics. 2005;21(16):3422–3.
NCBI latest BLAST executables [Available from: ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/. Accessed 30 November 2020].
Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36(Web Server issue):W181–4.
Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124.
Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28(9):1395–404.
Bray J. PubMLST: Public databases for molecular typing and microbial genome diversity - Ribosomal MLST. [Available from: https://pubmlst.org/species-id. Accessed 30 November 2020].
Meier-Kolthoff JPA, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based (sub-)species delineation: Genome-to-Genome Distance Calculator [Available from: http://ggdc.dsmz.de/ggdc.php. Accessed 30 November 2020].
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol. 2011;7:539.
Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–66.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4.
Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, et al. TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 2009;25(1):126–7.
Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61(6):1061–7.
Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–67.
Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009–10.
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–D32.
Acknowledgements
The authors would like to thank Dr. Frank Wright (BioSS, Edinburgh, UK), who sadly died at the end of 2020, for useful help and discussions regarding the phylogenetic and recombination analyses. He will be greatly missed.
Funding
This work was supported by funding to DL, ML and PR from the Scottish Government Rural and Environment Science and Analytical Services (RESAS) division; and to DSAB and DV through a post-doctoral grant from the Research Foundation Flanders (FWO-Vlaanderen).
Author information
Authors and Affiliations
Contributions
DV, YP and DL conceptualized the study; DB and DV conducted the genome sequencing and assembly of the raw data; AVDE and YP provided initial annotation of the sequence; PR conducted the recombination and whole genome phylogenetic analyses; DL and ML conducted the typing analyses, whole genome phylogenetic analyses of the chromosome and plasmid, and comparative genomic analyses, including analyses of virulence genes/regions; DL, ML and PR analysed and interpreted the data; DL wrote the manuscript with major contributions from ML and PR; all authors read and contributed to the draft, as well as reading and approving the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Results of rMLST analysis to determine species classification using the rps gene database at https://pubmlst.org/species-id.
Additional file 2: Fig. S1.
Minimum spanning tree illustrating cluster analysis of MLST sequence type (ST) profiles. Cluster analysis is based on nucleotide differences in seven MLST housekeeping gene fragments (enoA, fumC, gatA, gidA, hemN, hlfX and oppA). STs for strain C. psittaci strain 84/2334 and representative strains from Chlamydiaceae species (excluding C. trachomatis) are indicated (genotypes are given in square brackets within tree; numbers in square brackets in key indicate total strains included for each species).
Additional file 3: Fig. S2.
Phylogenetic tree of a 16S rRNA gene alignment of strain 84/2334 and other Chlamydiaceae species. The consensus tree for the 1470 bp alignment was estimated in TOPALi by Neighbour Joining using a F84 + G substitution and rate heterogeneity model and 100 non-parametric bootstrap replicates. The tree is midpoint rooted and bootstrap support is indicated by the number at the node. The scale bar indicates the expected substitutions per site. Genotypes are given in square brackets. The tree was prepared in Dendroscope. Strain 84/2334 is in bold and red font.
Additional file 4: Table S2.
Results of digital DNA-DNA hybridization to determine species using the Genome-to-Genome Distance Calculator 2.1 at http://ggdc.dsmz.de/ggdc.php.
Additional file 5: Fig. S3.
Whole genome phylogenetic analysis informed by recombination with added outgroup. Phylogenetic tree of a whole genome sequence MAFFT alignment of the C. abortus (Cab) and C. psittaci (Cps) strains shown in Table 2 as derived by Gubbins after removing genomic regions affected by recombination. Genotypes are given in square brackets. The sequence of C. pecorum W73 was additionally provided as outgroup. FastTree was used as tree builder, and a maximum of 100 iterations was specified in order to guarantee convergence. Tree was midpoint rooted and prepared using Dendroscope. Strain 84/2334 is in bold and red font. Classical and avian C. abortus strains are in blue and green fonts, respectively.
Additional file 6: Fig. S4.
Whole genome phylogenetic analysis. Phylogenetic tree of a whole genome sequence MAFFT alignment of the C. abortus (Cab) and C. psittaci (Cps) strains shown in Table 2, and C. buteonis strain RSHA. The consensus tree was estimated in IQ-Tree by Maximum Likelihood using a TVM + F + R2 substitution and rate heterogeneity model, according to BIC model selection, and 100 non-parametric bootstrap replicates. The tree is midpoint rooted and bootstrap support is indicated by the number at the node. The scale bar indicates the expected substitutions per site. Genotypes are given in square brackets. The tree was prepared in Dendroscope. Strain 84/2334 is in bold and red font. Classical and avian C. abortus strains are in blue and green fonts, respectively.
Additional file 7: Fig. S5.
Whole genome NeighborNet network analysis. Phylogenetic network of a whole genome sequence alignment of the C. abortus (Cab) and C. psittaci (Cps) strains shown in Table 2, using the NeighborNet distances transformation (Ordinary Least Squares variance and Lambda Frac of 1.0) and EqualAngle splits transformation. The scale bar indicates the expected substitutions per site. Genotypes of C. psittaci and avian C. abortus strains are indicated in brackets. The figure was generated using SplitsTree4. Strain 84/2334 is in bold and red font.
Additional file 8: Fig. S6.
Summary of Gubbins recombination analysis including avian C. abortus strains 15-59d/3 and 15-70d/24. The red blocks represent recombination events occurring on an internal branch of the phylogenetic tree, which are shared by several strains by common descent. The blue blocks indicate recombination events occurring on terminal branches of the phylogenetic tree, which are unique to a specific strain. The parameters used for the run are those described in Fig. 4.
Additional file 9: Fig. S7.
Comparative genome analysis of C. psittaci 84/2334. Whole genome comparisons of C. psittaci strain 84/2334 and (A) C. abortus strains S26/3, AB7 and LLG and (B) representative C. psittaci strains VS225, 84/55, GR9 and M56 depicting amino acid matches computed using Megablast blastn. Homology matches are indicated by the red vertical bars, while inverted matches (indicating areas of recombination) are coloured blue. Horizontal grey bars represent the forward and reverse strands of DNA with CDSs marked as arrows. The main regions of difference occur in two of the Pmp loci (circled areas 1 and 3), the PZ region (circled area 2) and TMH loci (circled area 4). Please note that the gaps shown for C. abortus strain LLG compared to 84/2334 in Pmp loci (circled areas 1 and 3) are due to the sequences not being complete in these regions rather than homology differences. The figure was generated using the Artemis Comparison Tool (ACT).
Additional file 10: Fig. S8
. Phylogenetic analysis of chlamydial cytotoxin predicted protein sequences. The consensus tree was estimated in IQ-Tree by Maximum Likelihood using a JTTDCMut + F + I + G4 substitution and rate heterogeneity model, according to BIC model selection, and 100 non-parametric bootstrap replicates. The tree is midpoint rooted and bootstrap support is indicated by the number at the node. The scale bar indicates the expected substitutions per site. The tree was prepared in Dendroscope. Strain 84/2334 is in bold and red font.
Additional file 11: Fig. S9
. Polymorphic membrane protein family phylogenies. Consensus trees for singleton PmpA, PmpB, PmpD and PmpH family members (A), for the PmpE family members (B) and PmpG family members (C) for strain 84/2334 and the C. abortus and C. psittaci strains/genotypes shown in Table 2 were estimated in IQ-Tree by Maximum Likelihood using substitution and rate heterogeneity models JTTDCMut + F + I + G4, JTT + F + R3 and JTT + F + R5, respectively, according to BIC model selection, and 100 non-parametric bootstrap replicates. The trees were midpoint rooted and bootstrap support is indicated by the number at the node (only values greater than 70 are shown). The scale bar indicates the expected substitutions per site. The trees were prepared in Dendroscope. Strain 84/2334 is in bold and red font. Classical and avian C. abortus strains are in blue and green fonts, respectively.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Longbottom, D., Livingstone, M., Ribeca, P. et al. Whole genome de novo sequencing and comparative genomic analyses suggests that Chlamydia psittaci strain 84/2334 should be reclassified as Chlamydia abortus species. BMC Genomics 22, 159 (2021). https://doi.org/10.1186/s12864-021-07477-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-07477-6